
Gammapapillomavirus 6
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Gammapapillomavirus
Average proteome isoelectric point is 5.83
Get precalculated fractions of proteins
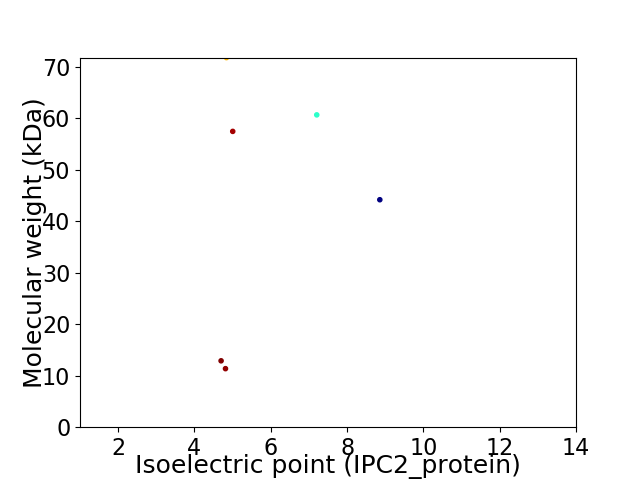
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
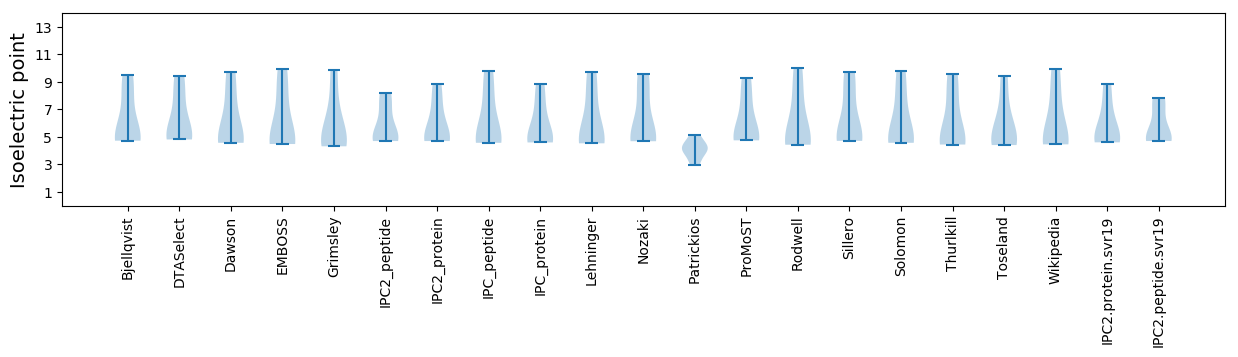
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2D2ALD5|A0A2D2ALD5_9PAPI Protein E7 OS=Gammapapillomavirus 6 OX=1175848 GN=E7 PE=3 SV=1
MM1 pKa = 7.99RR2 pKa = 11.84STEE5 pKa = 3.95PTIKK9 pKa = 10.53DD10 pKa = 3.03INVEE14 pKa = 4.1LEE16 pKa = 3.91PLVIPEE22 pKa = 4.25NLLCGEE28 pKa = 4.2VSLEE32 pKa = 4.02EE33 pKa = 4.11EE34 pKa = 4.3EE35 pKa = 4.96EE36 pKa = 4.2EE37 pKa = 5.09QEE39 pKa = 4.21EE40 pKa = 4.51QAQSPYY46 pKa = 10.79RR47 pKa = 11.84VVTQCKK53 pKa = 8.94HH54 pKa = 5.73CSKK57 pKa = 10.25QLCVYY62 pKa = 9.84VVATDD67 pKa = 3.0VGIRR71 pKa = 11.84IFEE74 pKa = 4.21HH75 pKa = 5.95VLCGDD80 pKa = 4.58LDD82 pKa = 4.51LVCPTCGRR90 pKa = 11.84QQHH93 pKa = 4.95RR94 pKa = 11.84HH95 pKa = 4.53GGQRR99 pKa = 11.84QQ100 pKa = 2.95
MM1 pKa = 7.99RR2 pKa = 11.84STEE5 pKa = 3.95PTIKK9 pKa = 10.53DD10 pKa = 3.03INVEE14 pKa = 4.1LEE16 pKa = 3.91PLVIPEE22 pKa = 4.25NLLCGEE28 pKa = 4.2VSLEE32 pKa = 4.02EE33 pKa = 4.11EE34 pKa = 4.3EE35 pKa = 4.96EE36 pKa = 4.2EE37 pKa = 5.09QEE39 pKa = 4.21EE40 pKa = 4.51QAQSPYY46 pKa = 10.79RR47 pKa = 11.84VVTQCKK53 pKa = 8.94HH54 pKa = 5.73CSKK57 pKa = 10.25QLCVYY62 pKa = 9.84VVATDD67 pKa = 3.0VGIRR71 pKa = 11.84IFEE74 pKa = 4.21HH75 pKa = 5.95VLCGDD80 pKa = 4.58LDD82 pKa = 4.51LVCPTCGRR90 pKa = 11.84QQHH93 pKa = 4.95RR94 pKa = 11.84HH95 pKa = 4.53GGQRR99 pKa = 11.84QQ100 pKa = 2.95
Molecular weight: 11.36 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2D2AL95|A0A2D2AL95_9PAPI E4 OS=Gammapapillomavirus 6 OX=1175848 GN=E4 PE=4 SV=1
MM1 pKa = 7.53EE2 pKa = 4.96NLVEE6 pKa = 5.3RR7 pKa = 11.84FDD9 pKa = 4.24AVQDD13 pKa = 3.82KK14 pKa = 10.77LLSLYY19 pKa = 10.19EE20 pKa = 4.39AGHH23 pKa = 6.57MDD25 pKa = 4.48LEE27 pKa = 4.49SQILHH32 pKa = 6.82WDD34 pKa = 3.83LEE36 pKa = 4.33RR37 pKa = 11.84QEE39 pKa = 5.0NVLLHH44 pKa = 6.22FARR47 pKa = 11.84KK48 pKa = 9.59QGIQNIGMQRR58 pKa = 11.84VPPLAATEE66 pKa = 4.25QKK68 pKa = 10.86AKK70 pKa = 8.82TAIYY74 pKa = 8.57MGLVLRR80 pKa = 11.84SLKK83 pKa = 10.26KK84 pKa = 10.18SPYY87 pKa = 9.36SQEE90 pKa = 3.51SWTLTEE96 pKa = 4.17TSNEE100 pKa = 4.05LFMTAPQNTFKK111 pKa = 11.0KK112 pKa = 9.63GGKK115 pKa = 7.76TIEE118 pKa = 3.85VLYY121 pKa = 11.05DD122 pKa = 3.51NEE124 pKa = 4.17EE125 pKa = 4.27TKK127 pKa = 11.15AMPYY131 pKa = 8.83TLWTAIYY138 pKa = 9.71YY139 pKa = 10.15QDD141 pKa = 5.39ADD143 pKa = 4.33DD144 pKa = 4.58MWHH147 pKa = 5.93KK148 pKa = 9.11TQGQVDD154 pKa = 4.28VKK156 pKa = 11.02GLFYY160 pKa = 11.25VDD162 pKa = 2.93ITGSKK167 pKa = 8.97TYY169 pKa = 10.99YY170 pKa = 10.56EE171 pKa = 5.4DD172 pKa = 3.5FAKK175 pKa = 10.58DD176 pKa = 3.1AKK178 pKa = 10.8KK179 pKa = 10.8YY180 pKa = 10.97GEE182 pKa = 4.03TGQWQVKK189 pKa = 8.19TGNDD193 pKa = 3.43TLFGPSIASTSEE205 pKa = 3.99DD206 pKa = 4.73NVDD209 pKa = 2.98GWQTIATPDD218 pKa = 3.0TGGPRR223 pKa = 11.84RR224 pKa = 11.84RR225 pKa = 11.84RR226 pKa = 11.84GRR228 pKa = 11.84PRR230 pKa = 11.84RR231 pKa = 11.84RR232 pKa = 11.84THH234 pKa = 6.32SATTSATSEE243 pKa = 4.16TDD245 pKa = 2.8TSSEE249 pKa = 3.7DD250 pKa = 2.95RR251 pKa = 11.84RR252 pKa = 11.84QRR254 pKa = 11.84RR255 pKa = 11.84GRR257 pKa = 11.84HH258 pKa = 3.97GKK260 pKa = 9.46RR261 pKa = 11.84RR262 pKa = 11.84SKK264 pKa = 10.23RR265 pKa = 11.84LRR267 pKa = 11.84WGSEE271 pKa = 3.42SSARR275 pKa = 11.84TPSPVGQGHH284 pKa = 6.95RR285 pKa = 11.84SPNKK289 pKa = 9.85HH290 pKa = 5.51YY291 pKa = 7.37PTRR294 pKa = 11.84LARR297 pKa = 11.84LQAEE301 pKa = 4.39AGDD304 pKa = 4.06PPVLLVKK311 pKa = 10.91GPANTTKK318 pKa = 9.94CWRR321 pKa = 11.84YY322 pKa = 9.14RR323 pKa = 11.84VKK325 pKa = 10.57LRR327 pKa = 11.84HH328 pKa = 6.56RR329 pKa = 11.84DD330 pKa = 3.35LFLCISTAFSWVGEE344 pKa = 4.38TGCDD348 pKa = 2.62RR349 pKa = 11.84VGCARR354 pKa = 11.84VLVGFVNEE362 pKa = 4.18TQRR365 pKa = 11.84EE366 pKa = 4.12RR367 pKa = 11.84FITTVRR373 pKa = 11.84LPKK376 pKa = 10.19GCTYY380 pKa = 11.38AFGNIDD386 pKa = 3.52SLL388 pKa = 4.29
MM1 pKa = 7.53EE2 pKa = 4.96NLVEE6 pKa = 5.3RR7 pKa = 11.84FDD9 pKa = 4.24AVQDD13 pKa = 3.82KK14 pKa = 10.77LLSLYY19 pKa = 10.19EE20 pKa = 4.39AGHH23 pKa = 6.57MDD25 pKa = 4.48LEE27 pKa = 4.49SQILHH32 pKa = 6.82WDD34 pKa = 3.83LEE36 pKa = 4.33RR37 pKa = 11.84QEE39 pKa = 5.0NVLLHH44 pKa = 6.22FARR47 pKa = 11.84KK48 pKa = 9.59QGIQNIGMQRR58 pKa = 11.84VPPLAATEE66 pKa = 4.25QKK68 pKa = 10.86AKK70 pKa = 8.82TAIYY74 pKa = 8.57MGLVLRR80 pKa = 11.84SLKK83 pKa = 10.26KK84 pKa = 10.18SPYY87 pKa = 9.36SQEE90 pKa = 3.51SWTLTEE96 pKa = 4.17TSNEE100 pKa = 4.05LFMTAPQNTFKK111 pKa = 11.0KK112 pKa = 9.63GGKK115 pKa = 7.76TIEE118 pKa = 3.85VLYY121 pKa = 11.05DD122 pKa = 3.51NEE124 pKa = 4.17EE125 pKa = 4.27TKK127 pKa = 11.15AMPYY131 pKa = 8.83TLWTAIYY138 pKa = 9.71YY139 pKa = 10.15QDD141 pKa = 5.39ADD143 pKa = 4.33DD144 pKa = 4.58MWHH147 pKa = 5.93KK148 pKa = 9.11TQGQVDD154 pKa = 4.28VKK156 pKa = 11.02GLFYY160 pKa = 11.25VDD162 pKa = 2.93ITGSKK167 pKa = 8.97TYY169 pKa = 10.99YY170 pKa = 10.56EE171 pKa = 5.4DD172 pKa = 3.5FAKK175 pKa = 10.58DD176 pKa = 3.1AKK178 pKa = 10.8KK179 pKa = 10.8YY180 pKa = 10.97GEE182 pKa = 4.03TGQWQVKK189 pKa = 8.19TGNDD193 pKa = 3.43TLFGPSIASTSEE205 pKa = 3.99DD206 pKa = 4.73NVDD209 pKa = 2.98GWQTIATPDD218 pKa = 3.0TGGPRR223 pKa = 11.84RR224 pKa = 11.84RR225 pKa = 11.84RR226 pKa = 11.84GRR228 pKa = 11.84PRR230 pKa = 11.84RR231 pKa = 11.84RR232 pKa = 11.84THH234 pKa = 6.32SATTSATSEE243 pKa = 4.16TDD245 pKa = 2.8TSSEE249 pKa = 3.7DD250 pKa = 2.95RR251 pKa = 11.84RR252 pKa = 11.84QRR254 pKa = 11.84RR255 pKa = 11.84GRR257 pKa = 11.84HH258 pKa = 3.97GKK260 pKa = 9.46RR261 pKa = 11.84RR262 pKa = 11.84SKK264 pKa = 10.23RR265 pKa = 11.84LRR267 pKa = 11.84WGSEE271 pKa = 3.42SSARR275 pKa = 11.84TPSPVGQGHH284 pKa = 6.95RR285 pKa = 11.84SPNKK289 pKa = 9.85HH290 pKa = 5.51YY291 pKa = 7.37PTRR294 pKa = 11.84LARR297 pKa = 11.84LQAEE301 pKa = 4.39AGDD304 pKa = 4.06PPVLLVKK311 pKa = 10.91GPANTTKK318 pKa = 9.94CWRR321 pKa = 11.84YY322 pKa = 9.14RR323 pKa = 11.84VKK325 pKa = 10.57LRR327 pKa = 11.84HH328 pKa = 6.56RR329 pKa = 11.84DD330 pKa = 3.35LFLCISTAFSWVGEE344 pKa = 4.38TGCDD348 pKa = 2.62RR349 pKa = 11.84VGCARR354 pKa = 11.84VLVGFVNEE362 pKa = 4.18TQRR365 pKa = 11.84EE366 pKa = 4.12RR367 pKa = 11.84FITTVRR373 pKa = 11.84LPKK376 pKa = 10.19GCTYY380 pKa = 11.38AFGNIDD386 pKa = 3.52SLL388 pKa = 4.29
Molecular weight: 44.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2317 |
100 |
634 |
386.2 |
43.03 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.388 ± 0.367 | 2.287 ± 0.733 |
6.647 ± 0.451 | 5.783 ± 0.78 |
3.712 ± 0.455 | 6.56 ± 0.781 |
1.813 ± 0.312 | 5.222 ± 0.768 |
4.963 ± 0.839 | 8.2 ± 0.796 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.813 ± 0.485 | 4.143 ± 0.676 |
6.388 ± 0.786 | 4.748 ± 0.403 |
5.913 ± 0.86 | 6.949 ± 0.426 |
7.035 ± 1.163 | 7.208 ± 0.684 |
1.252 ± 0.257 | 2.978 ± 0.275 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
