
Streptococcus satellite phage Javan309
Taxonomy: Viruses; unclassified bacterial viruses
Average proteome isoelectric point is 6.54
Get precalculated fractions of proteins
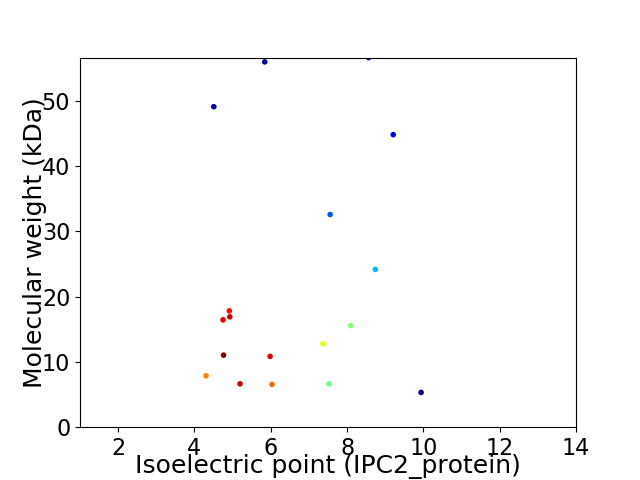
Virtual 2D-PAGE plot for 18 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4D5ZJA6|A0A4D5ZJA6_9VIRU Uncharacterized protein OS=Streptococcus satellite phage Javan309 OX=2558627 GN=JavanS309_0011 PE=4 SV=1
MM1 pKa = 7.74IYY3 pKa = 10.14SKK5 pKa = 10.8EE6 pKa = 3.99IVRR9 pKa = 11.84EE10 pKa = 3.93WLDD13 pKa = 3.73EE14 pKa = 4.04VAEE17 pKa = 4.16RR18 pKa = 11.84AKK20 pKa = 10.79DD21 pKa = 3.42HH22 pKa = 6.92PEE24 pKa = 3.57WVDD27 pKa = 3.29VFEE30 pKa = 4.78RR31 pKa = 11.84CYY33 pKa = 10.93TDD35 pKa = 3.42TLDD38 pKa = 3.54NTVEE42 pKa = 3.94ILEE45 pKa = 4.88DD46 pKa = 3.64GSTFVLTGDD55 pKa = 4.14IPAMWLRR62 pKa = 11.84DD63 pKa = 3.32STAQLRR69 pKa = 11.84PYY71 pKa = 10.38LHH73 pKa = 5.85VAKK76 pKa = 10.12RR77 pKa = 11.84DD78 pKa = 3.42PLLRR82 pKa = 11.84QTIAGLVKK90 pKa = 10.4RR91 pKa = 11.84QMTLILKK98 pKa = 10.09DD99 pKa = 3.66PYY101 pKa = 11.29ANSFNIEE108 pKa = 4.17EE109 pKa = 4.14NWKK112 pKa = 9.1GHH114 pKa = 6.42HH115 pKa = 5.69EE116 pKa = 4.16TDD118 pKa = 3.49HH119 pKa = 6.69TDD121 pKa = 3.16LNGWIWEE128 pKa = 4.15RR129 pKa = 11.84KK130 pKa = 9.0YY131 pKa = 10.94EE132 pKa = 4.16VDD134 pKa = 3.59SLCYY138 pKa = 9.51PLQLAYY144 pKa = 10.73LLWKK148 pKa = 8.5EE149 pKa = 4.32TGEE152 pKa = 4.0TSQFDD157 pKa = 3.91EE158 pKa = 4.66TFVAATKK165 pKa = 10.31EE166 pKa = 4.09ILHH169 pKa = 6.71LWTVEE174 pKa = 3.83QDD176 pKa = 3.86HH177 pKa = 6.81KK178 pKa = 10.74NSPYY182 pKa = 10.66RR183 pKa = 11.84FVRR186 pKa = 11.84DD187 pKa = 3.34TDD189 pKa = 3.58RR190 pKa = 11.84KK191 pKa = 10.6EE192 pKa = 3.82DD193 pKa = 3.61TLVNDD198 pKa = 4.14GFGPNFAVTGMTWSAFRR215 pKa = 11.84PSDD218 pKa = 3.51DD219 pKa = 3.59CCQYY223 pKa = 11.51SYY225 pKa = 11.01LIPSNMFAVVVLGYY239 pKa = 9.29VQEE242 pKa = 4.52IFAEE246 pKa = 4.29LDD248 pKa = 3.46LADD251 pKa = 4.14SQSVTTDD258 pKa = 2.75AKK260 pKa = 10.58RR261 pKa = 11.84LQSEE265 pKa = 3.97IQEE268 pKa = 4.64GIEE271 pKa = 3.9NYY273 pKa = 10.71AYY275 pKa = 9.5TSNSKK280 pKa = 10.75GEE282 pKa = 4.04KK283 pKa = 9.27IYY285 pKa = 11.05AFEE288 pKa = 4.42VDD290 pKa = 3.53GLGNASIMDD299 pKa = 4.24DD300 pKa = 3.79PNVPSLLAAPYY311 pKa = 10.21LGYY314 pKa = 10.7CSVDD318 pKa = 3.41DD319 pKa = 4.34EE320 pKa = 5.33VYY322 pKa = 9.84QATRR326 pKa = 11.84RR327 pKa = 11.84TILSPEE333 pKa = 3.71NPYY336 pKa = 10.25FYY338 pKa = 10.23QGEE341 pKa = 4.37YY342 pKa = 10.97ASGLGSSHH350 pKa = 5.8TFYY353 pKa = 10.98RR354 pKa = 11.84YY355 pKa = 8.88IWPIALSIQGLTTRR369 pKa = 11.84DD370 pKa = 3.3KK371 pKa = 11.55AEE373 pKa = 4.21KK374 pKa = 10.44KK375 pKa = 10.4FLLDD379 pKa = 3.92QLVACDD385 pKa = 3.54GGTGVMHH392 pKa = 7.19EE393 pKa = 4.79SFHH396 pKa = 8.56VDD398 pKa = 3.77DD399 pKa = 4.03PTLYY403 pKa = 10.06SRR405 pKa = 11.84EE406 pKa = 4.11WFSWANMMFCEE417 pKa = 5.34LVLDD421 pKa = 4.09YY422 pKa = 11.56LDD424 pKa = 3.63IRR426 pKa = 4.46
MM1 pKa = 7.74IYY3 pKa = 10.14SKK5 pKa = 10.8EE6 pKa = 3.99IVRR9 pKa = 11.84EE10 pKa = 3.93WLDD13 pKa = 3.73EE14 pKa = 4.04VAEE17 pKa = 4.16RR18 pKa = 11.84AKK20 pKa = 10.79DD21 pKa = 3.42HH22 pKa = 6.92PEE24 pKa = 3.57WVDD27 pKa = 3.29VFEE30 pKa = 4.78RR31 pKa = 11.84CYY33 pKa = 10.93TDD35 pKa = 3.42TLDD38 pKa = 3.54NTVEE42 pKa = 3.94ILEE45 pKa = 4.88DD46 pKa = 3.64GSTFVLTGDD55 pKa = 4.14IPAMWLRR62 pKa = 11.84DD63 pKa = 3.32STAQLRR69 pKa = 11.84PYY71 pKa = 10.38LHH73 pKa = 5.85VAKK76 pKa = 10.12RR77 pKa = 11.84DD78 pKa = 3.42PLLRR82 pKa = 11.84QTIAGLVKK90 pKa = 10.4RR91 pKa = 11.84QMTLILKK98 pKa = 10.09DD99 pKa = 3.66PYY101 pKa = 11.29ANSFNIEE108 pKa = 4.17EE109 pKa = 4.14NWKK112 pKa = 9.1GHH114 pKa = 6.42HH115 pKa = 5.69EE116 pKa = 4.16TDD118 pKa = 3.49HH119 pKa = 6.69TDD121 pKa = 3.16LNGWIWEE128 pKa = 4.15RR129 pKa = 11.84KK130 pKa = 9.0YY131 pKa = 10.94EE132 pKa = 4.16VDD134 pKa = 3.59SLCYY138 pKa = 9.51PLQLAYY144 pKa = 10.73LLWKK148 pKa = 8.5EE149 pKa = 4.32TGEE152 pKa = 4.0TSQFDD157 pKa = 3.91EE158 pKa = 4.66TFVAATKK165 pKa = 10.31EE166 pKa = 4.09ILHH169 pKa = 6.71LWTVEE174 pKa = 3.83QDD176 pKa = 3.86HH177 pKa = 6.81KK178 pKa = 10.74NSPYY182 pKa = 10.66RR183 pKa = 11.84FVRR186 pKa = 11.84DD187 pKa = 3.34TDD189 pKa = 3.58RR190 pKa = 11.84KK191 pKa = 10.6EE192 pKa = 3.82DD193 pKa = 3.61TLVNDD198 pKa = 4.14GFGPNFAVTGMTWSAFRR215 pKa = 11.84PSDD218 pKa = 3.51DD219 pKa = 3.59CCQYY223 pKa = 11.51SYY225 pKa = 11.01LIPSNMFAVVVLGYY239 pKa = 9.29VQEE242 pKa = 4.52IFAEE246 pKa = 4.29LDD248 pKa = 3.46LADD251 pKa = 4.14SQSVTTDD258 pKa = 2.75AKK260 pKa = 10.58RR261 pKa = 11.84LQSEE265 pKa = 3.97IQEE268 pKa = 4.64GIEE271 pKa = 3.9NYY273 pKa = 10.71AYY275 pKa = 9.5TSNSKK280 pKa = 10.75GEE282 pKa = 4.04KK283 pKa = 9.27IYY285 pKa = 11.05AFEE288 pKa = 4.42VDD290 pKa = 3.53GLGNASIMDD299 pKa = 4.24DD300 pKa = 3.79PNVPSLLAAPYY311 pKa = 10.21LGYY314 pKa = 10.7CSVDD318 pKa = 3.41DD319 pKa = 4.34EE320 pKa = 5.33VYY322 pKa = 9.84QATRR326 pKa = 11.84RR327 pKa = 11.84TILSPEE333 pKa = 3.71NPYY336 pKa = 10.25FYY338 pKa = 10.23QGEE341 pKa = 4.37YY342 pKa = 10.97ASGLGSSHH350 pKa = 5.8TFYY353 pKa = 10.98RR354 pKa = 11.84YY355 pKa = 8.88IWPIALSIQGLTTRR369 pKa = 11.84DD370 pKa = 3.3KK371 pKa = 11.55AEE373 pKa = 4.21KK374 pKa = 10.44KK375 pKa = 10.4FLLDD379 pKa = 3.92QLVACDD385 pKa = 3.54GGTGVMHH392 pKa = 7.19EE393 pKa = 4.79SFHH396 pKa = 8.56VDD398 pKa = 3.77DD399 pKa = 4.03PTLYY403 pKa = 10.06SRR405 pKa = 11.84EE406 pKa = 4.11WFSWANMMFCEE417 pKa = 5.34LVLDD421 pKa = 4.09YY422 pKa = 11.56LDD424 pKa = 3.63IRR426 pKa = 4.46
Molecular weight: 49.12 kDa
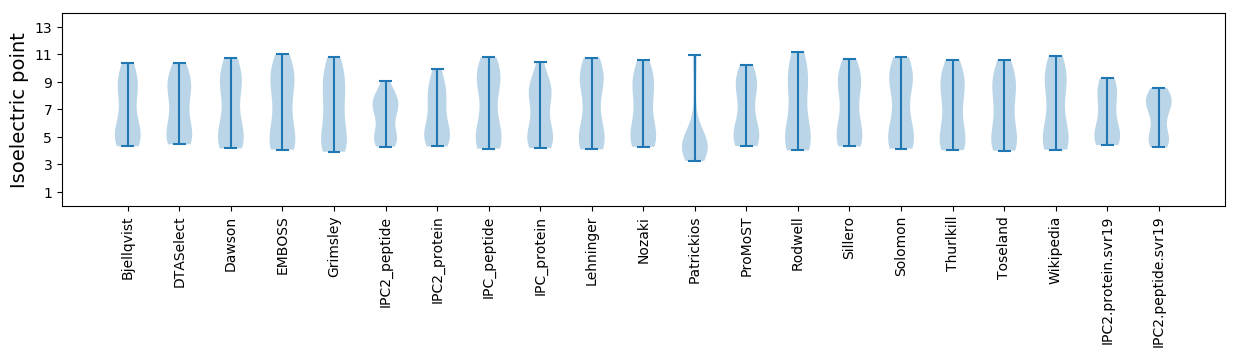
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4D5ZJ29|A0A4D5ZJ29_9VIRU Uncharacterized protein OS=Streptococcus satellite phage Javan309 OX=2558627 GN=JavanS309_0013 PE=4 SV=1
MM1 pKa = 7.56FDD3 pKa = 4.24SFPEE7 pKa = 3.89RR8 pKa = 11.84PKK10 pKa = 10.37YY11 pKa = 9.41MKK13 pKa = 9.51RR14 pKa = 11.84DD15 pKa = 3.72RR16 pKa = 11.84YY17 pKa = 9.01HH18 pKa = 4.93KK19 pKa = 10.08HH20 pKa = 4.07YY21 pKa = 10.89RR22 pKa = 11.84KK23 pKa = 9.39FLKK26 pKa = 9.0YY27 pKa = 8.31TKK29 pKa = 10.35KK30 pKa = 10.73GDD32 pKa = 3.79RR33 pKa = 11.84FWLNGSRR40 pKa = 11.84LL41 pKa = 3.57
MM1 pKa = 7.56FDD3 pKa = 4.24SFPEE7 pKa = 3.89RR8 pKa = 11.84PKK10 pKa = 10.37YY11 pKa = 9.41MKK13 pKa = 9.51RR14 pKa = 11.84DD15 pKa = 3.72RR16 pKa = 11.84YY17 pKa = 9.01HH18 pKa = 4.93KK19 pKa = 10.08HH20 pKa = 4.07YY21 pKa = 10.89RR22 pKa = 11.84KK23 pKa = 9.39FLKK26 pKa = 9.0YY27 pKa = 8.31TKK29 pKa = 10.35KK30 pKa = 10.73GDD32 pKa = 3.79RR33 pKa = 11.84FWLNGSRR40 pKa = 11.84LL41 pKa = 3.57
Molecular weight: 5.33 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3441 |
41 |
498 |
191.2 |
22.11 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.754 ± 0.248 | 0.639 ± 0.185 |
5.987 ± 0.494 | 8.253 ± 0.609 |
4.33 ± 0.222 | 5.202 ± 0.391 |
2.209 ± 0.255 | 6.51 ± 0.575 |
8.457 ± 0.659 | 9.59 ± 0.494 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.645 ± 0.291 | 4.999 ± 0.525 |
3.371 ± 0.463 | 3.981 ± 0.384 |
5.144 ± 0.341 | 5.812 ± 0.274 |
6.074 ± 0.452 | 5.086 ± 0.356 |
1.162 ± 0.228 | 4.795 ± 0.317 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
