
Chrysanthemum virus B (CVB)
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Tymovirales; Betaflexiviridae; Quinvirinae; Carlavirus
Average proteome isoelectric point is 6.87
Get precalculated fractions of proteins
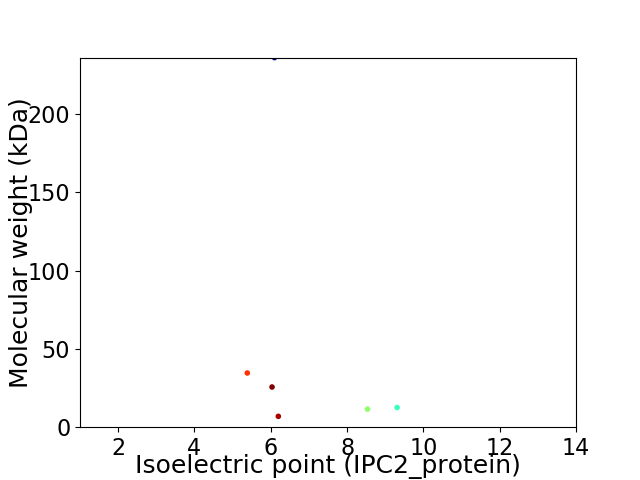
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q70HV5|Q70HV5_CVB Capsid protein OS=Chrysanthemum virus B OX=12165 GN=cp PE=3 SV=1
MM1 pKa = 7.59PPKK4 pKa = 9.63PAPGDD9 pKa = 3.68TEE11 pKa = 4.95GNASGSTPTPPPPPPAQTAEE31 pKa = 4.07EE32 pKa = 4.03ARR34 pKa = 11.84LRR36 pKa = 11.84LAEE39 pKa = 4.04MEE41 pKa = 4.14RR42 pKa = 11.84EE43 pKa = 3.96RR44 pKa = 11.84EE45 pKa = 3.96QEE47 pKa = 3.95QLLDD51 pKa = 3.89EE52 pKa = 4.78MNSNTPAEE60 pKa = 4.24DD61 pKa = 3.05ARR63 pKa = 11.84NISRR67 pKa = 11.84LTQLAALLRR76 pKa = 11.84RR77 pKa = 11.84EE78 pKa = 4.05QTNVHH83 pKa = 4.9VTNMALEE90 pKa = 4.31IGRR93 pKa = 11.84PALQPPPNMRR103 pKa = 11.84GDD105 pKa = 3.82PTNMYY110 pKa = 9.86SQVSTDD116 pKa = 4.22FLWKK120 pKa = 10.26IKK122 pKa = 8.41PQRR125 pKa = 11.84ISNNMATSEE134 pKa = 4.25DD135 pKa = 3.59MVKK138 pKa = 10.08IQVALEE144 pKa = 3.93GLGVPTEE151 pKa = 4.28SVKK154 pKa = 10.7EE155 pKa = 3.94VIIRR159 pKa = 11.84LVLNCANTSSSVYY172 pKa = 9.96QDD174 pKa = 3.22PKK176 pKa = 11.47GVIEE180 pKa = 4.22WDD182 pKa = 3.08GGAIIVDD189 pKa = 3.85DD190 pKa = 4.06VVGVITEE197 pKa = 4.13HH198 pKa = 4.79STLRR202 pKa = 11.84KK203 pKa = 9.18VCRR206 pKa = 11.84LYY208 pKa = 11.16AAVAWNYY215 pKa = 7.52MHH217 pKa = 7.29LQQTPPSDD225 pKa = 3.01WSAMGFHH232 pKa = 7.13PNVKK236 pKa = 9.69YY237 pKa = 10.54AAFDD241 pKa = 3.53FFDD244 pKa = 3.86YY245 pKa = 11.18VEE247 pKa = 4.49NGAAIRR253 pKa = 11.84PSGGIVPKK261 pKa = 8.44PTRR264 pKa = 11.84AEE266 pKa = 3.78YY267 pKa = 10.32VAYY270 pKa = 8.7NTYY273 pKa = 11.16KK274 pKa = 10.18MLALNKK280 pKa = 10.13ANNNDD285 pKa = 3.13TFGNFDD291 pKa = 3.47SAITGGRR298 pKa = 11.84QGPAIHH304 pKa = 6.74NNLNNANNKK313 pKa = 7.32TLL315 pKa = 3.5
MM1 pKa = 7.59PPKK4 pKa = 9.63PAPGDD9 pKa = 3.68TEE11 pKa = 4.95GNASGSTPTPPPPPPAQTAEE31 pKa = 4.07EE32 pKa = 4.03ARR34 pKa = 11.84LRR36 pKa = 11.84LAEE39 pKa = 4.04MEE41 pKa = 4.14RR42 pKa = 11.84EE43 pKa = 3.96RR44 pKa = 11.84EE45 pKa = 3.96QEE47 pKa = 3.95QLLDD51 pKa = 3.89EE52 pKa = 4.78MNSNTPAEE60 pKa = 4.24DD61 pKa = 3.05ARR63 pKa = 11.84NISRR67 pKa = 11.84LTQLAALLRR76 pKa = 11.84RR77 pKa = 11.84EE78 pKa = 4.05QTNVHH83 pKa = 4.9VTNMALEE90 pKa = 4.31IGRR93 pKa = 11.84PALQPPPNMRR103 pKa = 11.84GDD105 pKa = 3.82PTNMYY110 pKa = 9.86SQVSTDD116 pKa = 4.22FLWKK120 pKa = 10.26IKK122 pKa = 8.41PQRR125 pKa = 11.84ISNNMATSEE134 pKa = 4.25DD135 pKa = 3.59MVKK138 pKa = 10.08IQVALEE144 pKa = 3.93GLGVPTEE151 pKa = 4.28SVKK154 pKa = 10.7EE155 pKa = 3.94VIIRR159 pKa = 11.84LVLNCANTSSSVYY172 pKa = 9.96QDD174 pKa = 3.22PKK176 pKa = 11.47GVIEE180 pKa = 4.22WDD182 pKa = 3.08GGAIIVDD189 pKa = 3.85DD190 pKa = 4.06VVGVITEE197 pKa = 4.13HH198 pKa = 4.79STLRR202 pKa = 11.84KK203 pKa = 9.18VCRR206 pKa = 11.84LYY208 pKa = 11.16AAVAWNYY215 pKa = 7.52MHH217 pKa = 7.29LQQTPPSDD225 pKa = 3.01WSAMGFHH232 pKa = 7.13PNVKK236 pKa = 9.69YY237 pKa = 10.54AAFDD241 pKa = 3.53FFDD244 pKa = 3.86YY245 pKa = 11.18VEE247 pKa = 4.49NGAAIRR253 pKa = 11.84PSGGIVPKK261 pKa = 8.44PTRR264 pKa = 11.84AEE266 pKa = 3.78YY267 pKa = 10.32VAYY270 pKa = 8.7NTYY273 pKa = 11.16KK274 pKa = 10.18MLALNKK280 pKa = 10.13ANNNDD285 pKa = 3.13TFGNFDD291 pKa = 3.47SAITGGRR298 pKa = 11.84QGPAIHH304 pKa = 6.74NNLNNANNKK313 pKa = 7.32TLL315 pKa = 3.5
Molecular weight: 34.63 kDa
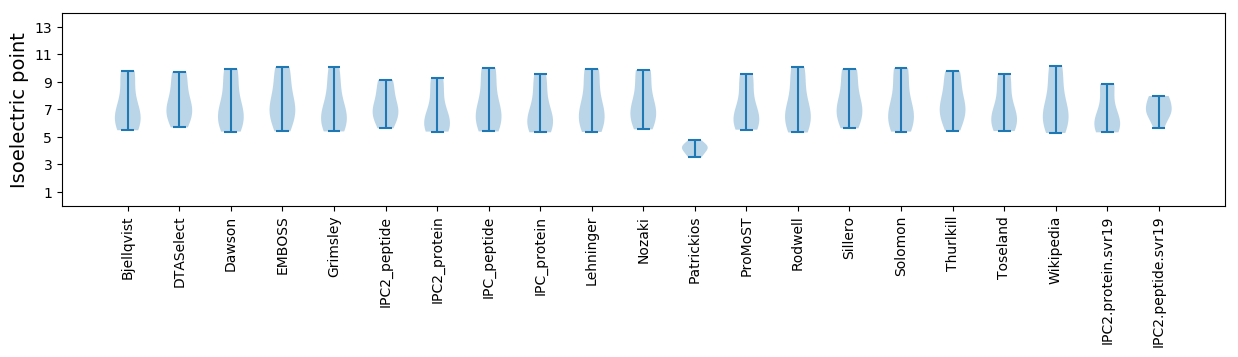
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q6ZZB7|Q6ZZB7_CVB 7 kDa protein OS=Chrysanthemum virus B OX=12165 GN=TGB3 PE=3 SV=1
MM1 pKa = 7.7DD2 pKa = 5.51VIVKK6 pKa = 8.46MLILRR11 pKa = 11.84KK12 pKa = 9.21FVEE15 pKa = 4.27QGNVCPIHH23 pKa = 6.7LCVDD27 pKa = 3.66IYY29 pKa = 11.39KK30 pKa = 10.33RR31 pKa = 11.84AFPRR35 pKa = 11.84SVNKK39 pKa = 10.0GRR41 pKa = 11.84SSYY44 pKa = 10.65ARR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84ALEE52 pKa = 3.44LGRR55 pKa = 11.84CHH57 pKa = 6.5RR58 pKa = 11.84CYY60 pKa = 10.09RR61 pKa = 11.84VYY63 pKa = 10.63PPLFPEE69 pKa = 5.31ISRR72 pKa = 11.84CDD74 pKa = 3.38NRR76 pKa = 11.84TCVPGISYY84 pKa = 10.39NSKK87 pKa = 9.44VRR89 pKa = 11.84DD90 pKa = 3.63YY91 pKa = 10.73ILWGVTEE98 pKa = 4.88VIPHH102 pKa = 6.76PGYY105 pKa = 11.18NFF107 pKa = 3.42
MM1 pKa = 7.7DD2 pKa = 5.51VIVKK6 pKa = 8.46MLILRR11 pKa = 11.84KK12 pKa = 9.21FVEE15 pKa = 4.27QGNVCPIHH23 pKa = 6.7LCVDD27 pKa = 3.66IYY29 pKa = 11.39KK30 pKa = 10.33RR31 pKa = 11.84AFPRR35 pKa = 11.84SVNKK39 pKa = 10.0GRR41 pKa = 11.84SSYY44 pKa = 10.65ARR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84ALEE52 pKa = 3.44LGRR55 pKa = 11.84CHH57 pKa = 6.5RR58 pKa = 11.84CYY60 pKa = 10.09RR61 pKa = 11.84VYY63 pKa = 10.63PPLFPEE69 pKa = 5.31ISRR72 pKa = 11.84CDD74 pKa = 3.38NRR76 pKa = 11.84TCVPGISYY84 pKa = 10.39NSKK87 pKa = 9.44VRR89 pKa = 11.84DD90 pKa = 3.63YY91 pKa = 10.73ILWGVTEE98 pKa = 4.88VIPHH102 pKa = 6.76PGYY105 pKa = 11.18NFF107 pKa = 3.42
Molecular weight: 12.58 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2906 |
64 |
2082 |
484.3 |
54.57 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.71 ± 0.683 | 2.753 ± 0.542 |
4.233 ± 0.218 | 7.983 ± 1.672 |
4.852 ± 0.781 | 7.054 ± 0.313 |
2.959 ± 0.331 | 4.852 ± 0.483 |
6.228 ± 1.237 | 9.601 ± 0.674 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.134 ± 0.365 | 4.783 ± 0.905 |
4.68 ± 1.353 | 2.719 ± 0.467 |
5.712 ± 0.746 | 6.641 ± 0.344 |
4.99 ± 0.565 | 6.435 ± 1.039 |
1.032 ± 0.239 | 3.648 ± 0.326 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
