
Wenling narna-like virus 4
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.97
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KIV8|A0A1L3KIV8_9VIRU RNA-dependent RNA polymerase OS=Wenling narna-like virus 4 OX=1923504 PE=4 SV=1
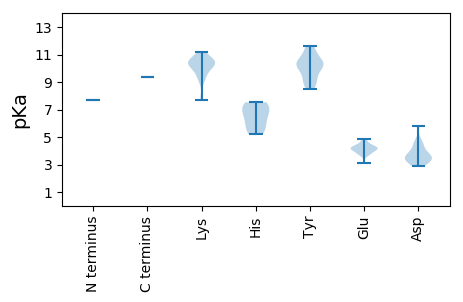
MM1 pKa = 7.69AIINGTDD8 pKa = 3.28RR9 pKa = 11.84LHH11 pKa = 6.74EE12 pKa = 4.88PYY14 pKa = 10.34NSPIVGEE21 pKa = 4.15LDD23 pKa = 3.49SVEE26 pKa = 4.85KK27 pKa = 10.47IITTMGIWARR37 pKa = 11.84GFNLNIDD44 pKa = 4.55HH45 pKa = 7.37ISPVCRR51 pKa = 11.84TFNSFKK57 pKa = 11.11DD58 pKa = 3.34LTTSIKK64 pKa = 9.89EE65 pKa = 3.75WSNYY69 pKa = 9.39LMNCLVSNKK78 pKa = 9.59VIRR81 pKa = 11.84PSIKK85 pKa = 10.24KK86 pKa = 9.13PFNRR90 pKa = 11.84NLYY93 pKa = 9.9DD94 pKa = 3.57YY95 pKa = 11.04LRR97 pKa = 11.84RR98 pKa = 11.84ACYY101 pKa = 10.22NPSNTQRR108 pKa = 11.84CMIISLLMVKK118 pKa = 10.06KK119 pKa = 10.36GFPEE123 pKa = 4.17APKK126 pKa = 10.67CLVDD130 pKa = 3.78EE131 pKa = 4.39TLKK134 pKa = 10.97DD135 pKa = 3.69HH136 pKa = 7.21QEE138 pKa = 4.17LLSSPSEE145 pKa = 4.02RR146 pKa = 11.84TDD148 pKa = 2.86EE149 pKa = 4.19KK150 pKa = 10.62TVDD153 pKa = 4.61DD154 pKa = 4.06ILEE157 pKa = 4.35IVDD160 pKa = 4.01KK161 pKa = 10.61LFPYY165 pKa = 10.44GWDD168 pKa = 3.53LNKK171 pKa = 10.59GICGPLPTKK180 pKa = 10.34AVYY183 pKa = 9.89EE184 pKa = 4.42GGVATQYY191 pKa = 10.43VFPEE195 pKa = 4.5EE196 pKa = 4.34ACSLIQQAKK205 pKa = 7.83LTYY208 pKa = 10.55DD209 pKa = 3.47RR210 pKa = 11.84FLHH213 pKa = 5.22MRR215 pKa = 11.84KK216 pKa = 9.02HH217 pKa = 5.82FKK219 pKa = 10.33VKK221 pKa = 10.23AVAVQQPGKK230 pKa = 10.2IRR232 pKa = 11.84VITKK236 pKa = 9.72SEE238 pKa = 4.02YY239 pKa = 10.34SLKK242 pKa = 10.73CLMPLQNALMDD253 pKa = 3.73QLRR256 pKa = 11.84NNPSFCLTRR265 pKa = 11.84KK266 pKa = 9.26EE267 pKa = 4.29CEE269 pKa = 4.09EE270 pKa = 3.94NDD272 pKa = 2.97FDD274 pKa = 5.84IFLEE278 pKa = 4.19EE279 pKa = 3.78KK280 pKa = 10.44GAFVSGDD287 pKa = 3.55YY288 pKa = 10.79KK289 pKa = 11.18SATDD293 pKa = 4.17NLNLDD298 pKa = 3.51LQYY301 pKa = 11.4YY302 pKa = 9.0IMQRR306 pKa = 11.84ILFNSVTPWVKK317 pKa = 10.55SLWMVALRR325 pKa = 11.84EE326 pKa = 4.17VSSHH330 pKa = 5.89KK331 pKa = 10.12IEE333 pKa = 4.06YY334 pKa = 8.78EE335 pKa = 3.81KK336 pKa = 10.88LQSIDD341 pKa = 3.37QVRR344 pKa = 11.84GQLMGSLLSFPLLCIINYY362 pKa = 10.22SIFHH366 pKa = 6.59ILFPNKK372 pKa = 9.55KK373 pKa = 9.76VLINGDD379 pKa = 4.27DD380 pKa = 3.37ILFRR384 pKa = 11.84ASQQEE389 pKa = 3.77YY390 pKa = 10.74DD391 pKa = 3.2LWKK394 pKa = 10.94VKK396 pKa = 10.39VLSVGLQLSIGKK408 pKa = 9.69NYY410 pKa = 9.85FSNEE414 pKa = 3.12IFTINSKK421 pKa = 10.12FYY423 pKa = 10.25RR424 pKa = 11.84WQDD427 pKa = 2.96RR428 pKa = 11.84ANEE431 pKa = 3.76IGYY434 pKa = 9.13LHH436 pKa = 7.27LGHH439 pKa = 7.52LLGPMSGDD447 pKa = 3.14SYY449 pKa = 11.39RR450 pKa = 11.84RR451 pKa = 11.84IPQRR455 pKa = 11.84FQGMYY460 pKa = 10.17KK461 pKa = 10.11RR462 pKa = 11.84YY463 pKa = 8.51CQKK466 pKa = 10.32PFEE469 pKa = 4.23NLRR472 pKa = 11.84DD473 pKa = 3.9LLEE476 pKa = 4.23PTCFGGLGGLTLDD489 pKa = 4.78EE490 pKa = 4.49YY491 pKa = 11.62NKK493 pKa = 9.21TVPVRR498 pKa = 11.84RR499 pKa = 11.84PSALAIHH506 pKa = 6.47HH507 pKa = 5.38QMKK510 pKa = 7.72TQEE513 pKa = 4.17VIKK516 pKa = 10.46LGSLAGDD523 pKa = 3.5VLDD526 pKa = 4.61FMSFKK531 pKa = 10.63FMTWIYY537 pKa = 8.78PSCRR541 pKa = 11.84RR542 pKa = 11.84EE543 pKa = 4.7DD544 pKa = 3.15IPQRR548 pKa = 11.84TRR550 pKa = 11.84IRR552 pKa = 11.84TKK554 pKa = 10.59GKK556 pKa = 10.16LGPLYY561 pKa = 9.36EE562 pKa = 4.06PRR564 pKa = 11.84MIEE567 pKa = 3.67RR568 pKa = 11.84TGNFMTFIGYY578 pKa = 9.34
MM1 pKa = 7.69AIINGTDD8 pKa = 3.28RR9 pKa = 11.84LHH11 pKa = 6.74EE12 pKa = 4.88PYY14 pKa = 10.34NSPIVGEE21 pKa = 4.15LDD23 pKa = 3.49SVEE26 pKa = 4.85KK27 pKa = 10.47IITTMGIWARR37 pKa = 11.84GFNLNIDD44 pKa = 4.55HH45 pKa = 7.37ISPVCRR51 pKa = 11.84TFNSFKK57 pKa = 11.11DD58 pKa = 3.34LTTSIKK64 pKa = 9.89EE65 pKa = 3.75WSNYY69 pKa = 9.39LMNCLVSNKK78 pKa = 9.59VIRR81 pKa = 11.84PSIKK85 pKa = 10.24KK86 pKa = 9.13PFNRR90 pKa = 11.84NLYY93 pKa = 9.9DD94 pKa = 3.57YY95 pKa = 11.04LRR97 pKa = 11.84RR98 pKa = 11.84ACYY101 pKa = 10.22NPSNTQRR108 pKa = 11.84CMIISLLMVKK118 pKa = 10.06KK119 pKa = 10.36GFPEE123 pKa = 4.17APKK126 pKa = 10.67CLVDD130 pKa = 3.78EE131 pKa = 4.39TLKK134 pKa = 10.97DD135 pKa = 3.69HH136 pKa = 7.21QEE138 pKa = 4.17LLSSPSEE145 pKa = 4.02RR146 pKa = 11.84TDD148 pKa = 2.86EE149 pKa = 4.19KK150 pKa = 10.62TVDD153 pKa = 4.61DD154 pKa = 4.06ILEE157 pKa = 4.35IVDD160 pKa = 4.01KK161 pKa = 10.61LFPYY165 pKa = 10.44GWDD168 pKa = 3.53LNKK171 pKa = 10.59GICGPLPTKK180 pKa = 10.34AVYY183 pKa = 9.89EE184 pKa = 4.42GGVATQYY191 pKa = 10.43VFPEE195 pKa = 4.5EE196 pKa = 4.34ACSLIQQAKK205 pKa = 7.83LTYY208 pKa = 10.55DD209 pKa = 3.47RR210 pKa = 11.84FLHH213 pKa = 5.22MRR215 pKa = 11.84KK216 pKa = 9.02HH217 pKa = 5.82FKK219 pKa = 10.33VKK221 pKa = 10.23AVAVQQPGKK230 pKa = 10.2IRR232 pKa = 11.84VITKK236 pKa = 9.72SEE238 pKa = 4.02YY239 pKa = 10.34SLKK242 pKa = 10.73CLMPLQNALMDD253 pKa = 3.73QLRR256 pKa = 11.84NNPSFCLTRR265 pKa = 11.84KK266 pKa = 9.26EE267 pKa = 4.29CEE269 pKa = 4.09EE270 pKa = 3.94NDD272 pKa = 2.97FDD274 pKa = 5.84IFLEE278 pKa = 4.19EE279 pKa = 3.78KK280 pKa = 10.44GAFVSGDD287 pKa = 3.55YY288 pKa = 10.79KK289 pKa = 11.18SATDD293 pKa = 4.17NLNLDD298 pKa = 3.51LQYY301 pKa = 11.4YY302 pKa = 9.0IMQRR306 pKa = 11.84ILFNSVTPWVKK317 pKa = 10.55SLWMVALRR325 pKa = 11.84EE326 pKa = 4.17VSSHH330 pKa = 5.89KK331 pKa = 10.12IEE333 pKa = 4.06YY334 pKa = 8.78EE335 pKa = 3.81KK336 pKa = 10.88LQSIDD341 pKa = 3.37QVRR344 pKa = 11.84GQLMGSLLSFPLLCIINYY362 pKa = 10.22SIFHH366 pKa = 6.59ILFPNKK372 pKa = 9.55KK373 pKa = 9.76VLINGDD379 pKa = 4.27DD380 pKa = 3.37ILFRR384 pKa = 11.84ASQQEE389 pKa = 3.77YY390 pKa = 10.74DD391 pKa = 3.2LWKK394 pKa = 10.94VKK396 pKa = 10.39VLSVGLQLSIGKK408 pKa = 9.69NYY410 pKa = 9.85FSNEE414 pKa = 3.12IFTINSKK421 pKa = 10.12FYY423 pKa = 10.25RR424 pKa = 11.84WQDD427 pKa = 2.96RR428 pKa = 11.84ANEE431 pKa = 3.76IGYY434 pKa = 9.13LHH436 pKa = 7.27LGHH439 pKa = 7.52LLGPMSGDD447 pKa = 3.14SYY449 pKa = 11.39RR450 pKa = 11.84RR451 pKa = 11.84IPQRR455 pKa = 11.84FQGMYY460 pKa = 10.17KK461 pKa = 10.11RR462 pKa = 11.84YY463 pKa = 8.51CQKK466 pKa = 10.32PFEE469 pKa = 4.23NLRR472 pKa = 11.84DD473 pKa = 3.9LLEE476 pKa = 4.23PTCFGGLGGLTLDD489 pKa = 4.78EE490 pKa = 4.49YY491 pKa = 11.62NKK493 pKa = 9.21TVPVRR498 pKa = 11.84RR499 pKa = 11.84PSALAIHH506 pKa = 6.47HH507 pKa = 5.38QMKK510 pKa = 7.72TQEE513 pKa = 4.17VIKK516 pKa = 10.46LGSLAGDD523 pKa = 3.5VLDD526 pKa = 4.61FMSFKK531 pKa = 10.63FMTWIYY537 pKa = 8.78PSCRR541 pKa = 11.84RR542 pKa = 11.84EE543 pKa = 4.7DD544 pKa = 3.15IPQRR548 pKa = 11.84TRR550 pKa = 11.84IRR552 pKa = 11.84TKK554 pKa = 10.59GKK556 pKa = 10.16LGPLYY561 pKa = 9.36EE562 pKa = 4.06PRR564 pKa = 11.84MIEE567 pKa = 3.67RR568 pKa = 11.84TGNFMTFIGYY578 pKa = 9.34
Molecular weight: 66.92 kDa
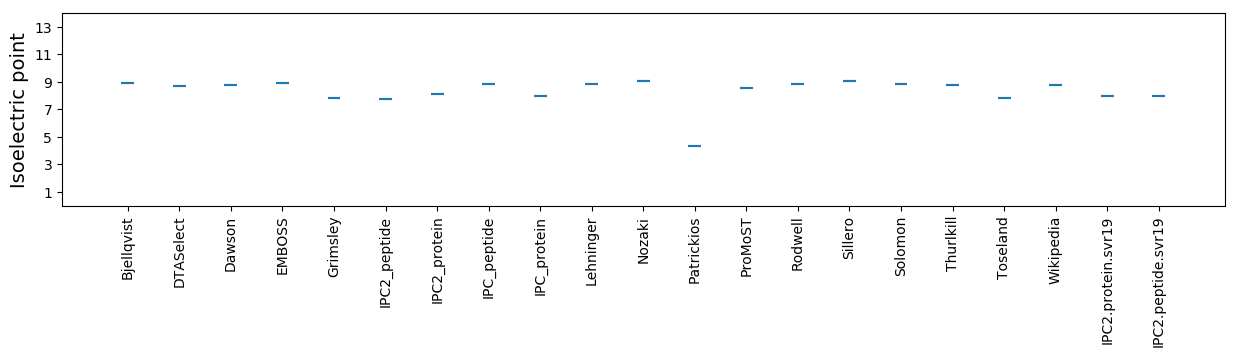
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KIV8|A0A1L3KIV8_9VIRU RNA-dependent RNA polymerase OS=Wenling narna-like virus 4 OX=1923504 PE=4 SV=1
MM1 pKa = 7.69AIINGTDD8 pKa = 3.28RR9 pKa = 11.84LHH11 pKa = 6.74EE12 pKa = 4.88PYY14 pKa = 10.34NSPIVGEE21 pKa = 4.15LDD23 pKa = 3.49SVEE26 pKa = 4.85KK27 pKa = 10.47IITTMGIWARR37 pKa = 11.84GFNLNIDD44 pKa = 4.55HH45 pKa = 7.37ISPVCRR51 pKa = 11.84TFNSFKK57 pKa = 11.11DD58 pKa = 3.34LTTSIKK64 pKa = 9.89EE65 pKa = 3.75WSNYY69 pKa = 9.39LMNCLVSNKK78 pKa = 9.59VIRR81 pKa = 11.84PSIKK85 pKa = 10.24KK86 pKa = 9.13PFNRR90 pKa = 11.84NLYY93 pKa = 9.9DD94 pKa = 3.57YY95 pKa = 11.04LRR97 pKa = 11.84RR98 pKa = 11.84ACYY101 pKa = 10.22NPSNTQRR108 pKa = 11.84CMIISLLMVKK118 pKa = 10.06KK119 pKa = 10.36GFPEE123 pKa = 4.17APKK126 pKa = 10.67CLVDD130 pKa = 3.78EE131 pKa = 4.39TLKK134 pKa = 10.97DD135 pKa = 3.69HH136 pKa = 7.21QEE138 pKa = 4.17LLSSPSEE145 pKa = 4.02RR146 pKa = 11.84TDD148 pKa = 2.86EE149 pKa = 4.19KK150 pKa = 10.62TVDD153 pKa = 4.61DD154 pKa = 4.06ILEE157 pKa = 4.35IVDD160 pKa = 4.01KK161 pKa = 10.61LFPYY165 pKa = 10.44GWDD168 pKa = 3.53LNKK171 pKa = 10.59GICGPLPTKK180 pKa = 10.34AVYY183 pKa = 9.89EE184 pKa = 4.42GGVATQYY191 pKa = 10.43VFPEE195 pKa = 4.5EE196 pKa = 4.34ACSLIQQAKK205 pKa = 7.83LTYY208 pKa = 10.55DD209 pKa = 3.47RR210 pKa = 11.84FLHH213 pKa = 5.22MRR215 pKa = 11.84KK216 pKa = 9.02HH217 pKa = 5.82FKK219 pKa = 10.33VKK221 pKa = 10.23AVAVQQPGKK230 pKa = 10.2IRR232 pKa = 11.84VITKK236 pKa = 9.72SEE238 pKa = 4.02YY239 pKa = 10.34SLKK242 pKa = 10.73CLMPLQNALMDD253 pKa = 3.73QLRR256 pKa = 11.84NNPSFCLTRR265 pKa = 11.84KK266 pKa = 9.26EE267 pKa = 4.29CEE269 pKa = 4.09EE270 pKa = 3.94NDD272 pKa = 2.97FDD274 pKa = 5.84IFLEE278 pKa = 4.19EE279 pKa = 3.78KK280 pKa = 10.44GAFVSGDD287 pKa = 3.55YY288 pKa = 10.79KK289 pKa = 11.18SATDD293 pKa = 4.17NLNLDD298 pKa = 3.51LQYY301 pKa = 11.4YY302 pKa = 9.0IMQRR306 pKa = 11.84ILFNSVTPWVKK317 pKa = 10.55SLWMVALRR325 pKa = 11.84EE326 pKa = 4.17VSSHH330 pKa = 5.89KK331 pKa = 10.12IEE333 pKa = 4.06YY334 pKa = 8.78EE335 pKa = 3.81KK336 pKa = 10.88LQSIDD341 pKa = 3.37QVRR344 pKa = 11.84GQLMGSLLSFPLLCIINYY362 pKa = 10.22SIFHH366 pKa = 6.59ILFPNKK372 pKa = 9.55KK373 pKa = 9.76VLINGDD379 pKa = 4.27DD380 pKa = 3.37ILFRR384 pKa = 11.84ASQQEE389 pKa = 3.77YY390 pKa = 10.74DD391 pKa = 3.2LWKK394 pKa = 10.94VKK396 pKa = 10.39VLSVGLQLSIGKK408 pKa = 9.69NYY410 pKa = 9.85FSNEE414 pKa = 3.12IFTINSKK421 pKa = 10.12FYY423 pKa = 10.25RR424 pKa = 11.84WQDD427 pKa = 2.96RR428 pKa = 11.84ANEE431 pKa = 3.76IGYY434 pKa = 9.13LHH436 pKa = 7.27LGHH439 pKa = 7.52LLGPMSGDD447 pKa = 3.14SYY449 pKa = 11.39RR450 pKa = 11.84RR451 pKa = 11.84IPQRR455 pKa = 11.84FQGMYY460 pKa = 10.17KK461 pKa = 10.11RR462 pKa = 11.84YY463 pKa = 8.51CQKK466 pKa = 10.32PFEE469 pKa = 4.23NLRR472 pKa = 11.84DD473 pKa = 3.9LLEE476 pKa = 4.23PTCFGGLGGLTLDD489 pKa = 4.78EE490 pKa = 4.49YY491 pKa = 11.62NKK493 pKa = 9.21TVPVRR498 pKa = 11.84RR499 pKa = 11.84PSALAIHH506 pKa = 6.47HH507 pKa = 5.38QMKK510 pKa = 7.72TQEE513 pKa = 4.17VIKK516 pKa = 10.46LGSLAGDD523 pKa = 3.5VLDD526 pKa = 4.61FMSFKK531 pKa = 10.63FMTWIYY537 pKa = 8.78PSCRR541 pKa = 11.84RR542 pKa = 11.84EE543 pKa = 4.7DD544 pKa = 3.15IPQRR548 pKa = 11.84TRR550 pKa = 11.84IRR552 pKa = 11.84TKK554 pKa = 10.59GKK556 pKa = 10.16LGPLYY561 pKa = 9.36EE562 pKa = 4.06PRR564 pKa = 11.84MIEE567 pKa = 3.67RR568 pKa = 11.84TGNFMTFIGYY578 pKa = 9.34
MM1 pKa = 7.69AIINGTDD8 pKa = 3.28RR9 pKa = 11.84LHH11 pKa = 6.74EE12 pKa = 4.88PYY14 pKa = 10.34NSPIVGEE21 pKa = 4.15LDD23 pKa = 3.49SVEE26 pKa = 4.85KK27 pKa = 10.47IITTMGIWARR37 pKa = 11.84GFNLNIDD44 pKa = 4.55HH45 pKa = 7.37ISPVCRR51 pKa = 11.84TFNSFKK57 pKa = 11.11DD58 pKa = 3.34LTTSIKK64 pKa = 9.89EE65 pKa = 3.75WSNYY69 pKa = 9.39LMNCLVSNKK78 pKa = 9.59VIRR81 pKa = 11.84PSIKK85 pKa = 10.24KK86 pKa = 9.13PFNRR90 pKa = 11.84NLYY93 pKa = 9.9DD94 pKa = 3.57YY95 pKa = 11.04LRR97 pKa = 11.84RR98 pKa = 11.84ACYY101 pKa = 10.22NPSNTQRR108 pKa = 11.84CMIISLLMVKK118 pKa = 10.06KK119 pKa = 10.36GFPEE123 pKa = 4.17APKK126 pKa = 10.67CLVDD130 pKa = 3.78EE131 pKa = 4.39TLKK134 pKa = 10.97DD135 pKa = 3.69HH136 pKa = 7.21QEE138 pKa = 4.17LLSSPSEE145 pKa = 4.02RR146 pKa = 11.84TDD148 pKa = 2.86EE149 pKa = 4.19KK150 pKa = 10.62TVDD153 pKa = 4.61DD154 pKa = 4.06ILEE157 pKa = 4.35IVDD160 pKa = 4.01KK161 pKa = 10.61LFPYY165 pKa = 10.44GWDD168 pKa = 3.53LNKK171 pKa = 10.59GICGPLPTKK180 pKa = 10.34AVYY183 pKa = 9.89EE184 pKa = 4.42GGVATQYY191 pKa = 10.43VFPEE195 pKa = 4.5EE196 pKa = 4.34ACSLIQQAKK205 pKa = 7.83LTYY208 pKa = 10.55DD209 pKa = 3.47RR210 pKa = 11.84FLHH213 pKa = 5.22MRR215 pKa = 11.84KK216 pKa = 9.02HH217 pKa = 5.82FKK219 pKa = 10.33VKK221 pKa = 10.23AVAVQQPGKK230 pKa = 10.2IRR232 pKa = 11.84VITKK236 pKa = 9.72SEE238 pKa = 4.02YY239 pKa = 10.34SLKK242 pKa = 10.73CLMPLQNALMDD253 pKa = 3.73QLRR256 pKa = 11.84NNPSFCLTRR265 pKa = 11.84KK266 pKa = 9.26EE267 pKa = 4.29CEE269 pKa = 4.09EE270 pKa = 3.94NDD272 pKa = 2.97FDD274 pKa = 5.84IFLEE278 pKa = 4.19EE279 pKa = 3.78KK280 pKa = 10.44GAFVSGDD287 pKa = 3.55YY288 pKa = 10.79KK289 pKa = 11.18SATDD293 pKa = 4.17NLNLDD298 pKa = 3.51LQYY301 pKa = 11.4YY302 pKa = 9.0IMQRR306 pKa = 11.84ILFNSVTPWVKK317 pKa = 10.55SLWMVALRR325 pKa = 11.84EE326 pKa = 4.17VSSHH330 pKa = 5.89KK331 pKa = 10.12IEE333 pKa = 4.06YY334 pKa = 8.78EE335 pKa = 3.81KK336 pKa = 10.88LQSIDD341 pKa = 3.37QVRR344 pKa = 11.84GQLMGSLLSFPLLCIINYY362 pKa = 10.22SIFHH366 pKa = 6.59ILFPNKK372 pKa = 9.55KK373 pKa = 9.76VLINGDD379 pKa = 4.27DD380 pKa = 3.37ILFRR384 pKa = 11.84ASQQEE389 pKa = 3.77YY390 pKa = 10.74DD391 pKa = 3.2LWKK394 pKa = 10.94VKK396 pKa = 10.39VLSVGLQLSIGKK408 pKa = 9.69NYY410 pKa = 9.85FSNEE414 pKa = 3.12IFTINSKK421 pKa = 10.12FYY423 pKa = 10.25RR424 pKa = 11.84WQDD427 pKa = 2.96RR428 pKa = 11.84ANEE431 pKa = 3.76IGYY434 pKa = 9.13LHH436 pKa = 7.27LGHH439 pKa = 7.52LLGPMSGDD447 pKa = 3.14SYY449 pKa = 11.39RR450 pKa = 11.84RR451 pKa = 11.84IPQRR455 pKa = 11.84FQGMYY460 pKa = 10.17KK461 pKa = 10.11RR462 pKa = 11.84YY463 pKa = 8.51CQKK466 pKa = 10.32PFEE469 pKa = 4.23NLRR472 pKa = 11.84DD473 pKa = 3.9LLEE476 pKa = 4.23PTCFGGLGGLTLDD489 pKa = 4.78EE490 pKa = 4.49YY491 pKa = 11.62NKK493 pKa = 9.21TVPVRR498 pKa = 11.84RR499 pKa = 11.84PSALAIHH506 pKa = 6.47HH507 pKa = 5.38QMKK510 pKa = 7.72TQEE513 pKa = 4.17VIKK516 pKa = 10.46LGSLAGDD523 pKa = 3.5VLDD526 pKa = 4.61FMSFKK531 pKa = 10.63FMTWIYY537 pKa = 8.78PSCRR541 pKa = 11.84RR542 pKa = 11.84EE543 pKa = 4.7DD544 pKa = 3.15IPQRR548 pKa = 11.84TRR550 pKa = 11.84IRR552 pKa = 11.84TKK554 pKa = 10.59GKK556 pKa = 10.16LGPLYY561 pKa = 9.36EE562 pKa = 4.06PRR564 pKa = 11.84MIEE567 pKa = 3.67RR568 pKa = 11.84TGNFMTFIGYY578 pKa = 9.34
Molecular weight: 66.92 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
578 |
578 |
578 |
578.0 |
66.92 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.287 ± 0.0 | 2.422 ± 0.0 |
5.19 ± 0.0 | 5.536 ± 0.0 |
5.017 ± 0.0 | 5.709 ± 0.0 |
1.903 ± 0.0 | 7.266 ± 0.0 |
7.093 ± 0.0 | 10.9 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.114 ± 0.0 | 5.19 ± 0.0 |
5.017 ± 0.0 | 4.152 ± 0.0 |
5.882 ± 0.0 | 6.574 ± 0.0 |
4.671 ± 0.0 | 5.19 ± 0.0 |
1.384 ± 0.0 | 4.498 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
