
Desulfobacteraceae bacterium SEEP-SAG9
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Desulfobacterales; Desulfobacteraceae; unclassified Desulfobacteraceae
Average proteome isoelectric point is 6.89
Get precalculated fractions of proteins
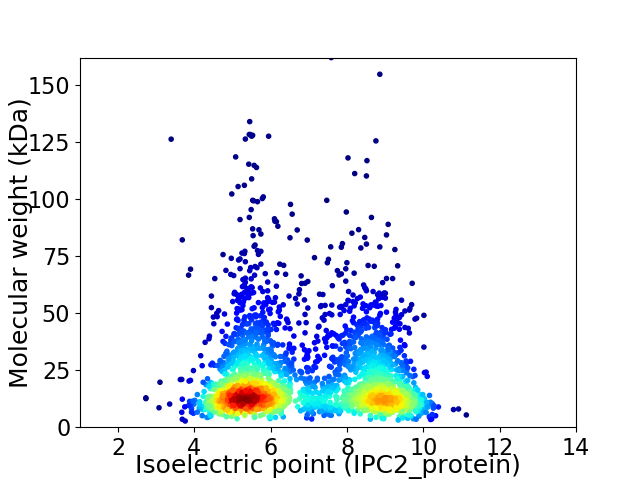
Virtual 2D-PAGE plot for 2621 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2S8K856|A0A2S8K856_9DELT GMC_oxred_C domain-containing protein OS=Desulfobacteraceae bacterium SEEP-SAG9 OX=2100161 GN=C6A37_03145 PE=4 SV=1
MM1 pKa = 7.27KK2 pKa = 9.68TRR4 pKa = 11.84LVINMLFALLFFISGVSAQTTYY26 pKa = 10.54IVAPSGGDD34 pKa = 3.09FTTIQDD40 pKa = 4.79CINGAINGDD49 pKa = 3.17ICLISPGVYY58 pKa = 9.71VEE60 pKa = 4.91NIDD63 pKa = 5.04FIGKK67 pKa = 8.71NLTVKK72 pKa = 10.53SSSGPEE78 pKa = 3.5ATSIDD83 pKa = 3.96GADD86 pKa = 3.43SGSVVRR92 pKa = 11.84FQTNEE97 pKa = 3.53TAEE100 pKa = 4.74AILDD104 pKa = 3.5GFMIRR109 pKa = 11.84NGNDD113 pKa = 3.15LQGGGIFCQGSSPTIKK129 pKa = 10.58NCIITEE135 pKa = 4.03NEE137 pKa = 4.02VYY139 pKa = 10.59GIEE142 pKa = 4.34GSDD145 pKa = 3.72GEE147 pKa = 4.8GAGIYY152 pKa = 10.05CNNSSPRR159 pKa = 11.84IMDD162 pKa = 3.23CTISYY167 pKa = 10.58NIAAVSDD174 pKa = 3.53YY175 pKa = 10.93GYY177 pKa = 11.04AYY179 pKa = 10.46AHH181 pKa = 6.78GGGIYY186 pKa = 10.11SVQSSSTFTDD196 pKa = 3.43CTITHH201 pKa = 6.47NEE203 pKa = 3.62AGTFGGGISCWDD215 pKa = 3.49SSSSTFTDD223 pKa = 4.75CIISNNSALGRR234 pKa = 11.84GGGGIYY240 pKa = 10.28SSSTLMIEE248 pKa = 4.01RR249 pKa = 11.84CTVSDD254 pKa = 3.38NYY256 pKa = 10.87AYY258 pKa = 10.28QGGGIYY264 pKa = 9.99SYY266 pKa = 11.91GEE268 pKa = 4.0TTISDD273 pKa = 4.01SIIAGNSSDD282 pKa = 4.83DD283 pKa = 3.76DD284 pKa = 4.37GGGIFSYY291 pKa = 11.38GNILSLVNCTIFNNSGNVGGGVCFKK316 pKa = 10.6GDD318 pKa = 3.99CNATIANSIIADD330 pKa = 3.53NTGNCGGGIAVWGSPSGPGSFSITEE355 pKa = 4.07STISGNTASGCGGGFWFDD373 pKa = 3.75YY374 pKa = 11.05VPLATVTNCMIFRR387 pKa = 11.84NTALGGGGGGGNITGEE403 pKa = 4.37TSTMINSSTFFGNSSTEE420 pKa = 3.72VGGGIFYY427 pKa = 8.93RR428 pKa = 11.84TDD430 pKa = 3.32GDD432 pKa = 3.8NVSSFNVIKK441 pKa = 10.91NCILWDD447 pKa = 4.08DD448 pKa = 4.13SATDD452 pKa = 4.04GPEE455 pKa = 3.42IALVEE460 pKa = 4.37EE461 pKa = 4.64YY462 pKa = 10.96GDD464 pKa = 4.03PEE466 pKa = 4.62LSVEE470 pKa = 4.05MSYY473 pKa = 11.72SDD475 pKa = 3.63VKK477 pKa = 11.0GGQSDD482 pKa = 3.33VHH484 pKa = 8.22IIGGVLNWQAGNIEE498 pKa = 4.64SNPLFVDD505 pKa = 3.59PDD507 pKa = 3.94GDD509 pKa = 4.32DD510 pKa = 3.99NLPGTLDD517 pKa = 4.19DD518 pKa = 5.56DD519 pKa = 4.24FHH521 pKa = 6.42LTSDD525 pKa = 4.38SLCIDD530 pKa = 4.01AGTSQDD536 pKa = 4.01APSIDD541 pKa = 3.27IDD543 pKa = 4.19GDD545 pKa = 3.68PRR547 pKa = 11.84PQGAAIDD554 pKa = 3.96IGADD558 pKa = 3.52EE559 pKa = 5.38YY560 pKa = 10.98IDD562 pKa = 4.6PDD564 pKa = 3.66SDD566 pKa = 3.85GDD568 pKa = 4.14GIEE571 pKa = 4.06YY572 pKa = 10.55GIDD575 pKa = 3.06IDD577 pKa = 3.85PTVFSSEE584 pKa = 3.92FSDD587 pKa = 5.25AEE589 pKa = 4.09TTYY592 pKa = 11.22GIIIDD597 pKa = 4.48RR598 pKa = 11.84GDD600 pKa = 3.45QVVTVIDD607 pKa = 4.18LPDD610 pKa = 4.37PNGVRR615 pKa = 11.84IAASGGDD622 pKa = 3.65TPASVSACGGSAIFTLNDD640 pKa = 3.08GDD642 pKa = 5.12VITFTCGSVKK652 pKa = 10.04IEE654 pKa = 4.34VISGPVKK661 pKa = 10.24ISFVADD667 pKa = 3.89DD668 pKa = 4.15GTTATTTLDD677 pKa = 3.26AGNILTFEE685 pKa = 4.7PEE687 pKa = 4.15TLIITAPDD695 pKa = 3.64TNTEE699 pKa = 3.85NAVIVVDD706 pKa = 4.21GKK708 pKa = 11.06EE709 pKa = 3.81FAIEE713 pKa = 3.99PGEE716 pKa = 4.24SVEE719 pKa = 4.27LDD721 pKa = 2.98ITIVIDD727 pKa = 4.07GCDD730 pKa = 3.46TGVLDD735 pKa = 6.02SDD737 pKa = 5.11YY738 pKa = 10.45EE739 pKa = 4.14DD740 pKa = 3.71QSISEE745 pKa = 4.77WISQCAEE752 pKa = 3.49NAKK755 pKa = 10.02NHH757 pKa = 5.21GKK759 pKa = 10.04YY760 pKa = 10.01VSCVAKK766 pKa = 9.27LTKK769 pKa = 9.88KK770 pKa = 10.12LKK772 pKa = 9.47KK773 pKa = 10.43AGVITGKK780 pKa = 10.4EE781 pKa = 3.48KK782 pKa = 10.98GAIQSCAAQADD793 pKa = 3.61IHH795 pKa = 6.3
MM1 pKa = 7.27KK2 pKa = 9.68TRR4 pKa = 11.84LVINMLFALLFFISGVSAQTTYY26 pKa = 10.54IVAPSGGDD34 pKa = 3.09FTTIQDD40 pKa = 4.79CINGAINGDD49 pKa = 3.17ICLISPGVYY58 pKa = 9.71VEE60 pKa = 4.91NIDD63 pKa = 5.04FIGKK67 pKa = 8.71NLTVKK72 pKa = 10.53SSSGPEE78 pKa = 3.5ATSIDD83 pKa = 3.96GADD86 pKa = 3.43SGSVVRR92 pKa = 11.84FQTNEE97 pKa = 3.53TAEE100 pKa = 4.74AILDD104 pKa = 3.5GFMIRR109 pKa = 11.84NGNDD113 pKa = 3.15LQGGGIFCQGSSPTIKK129 pKa = 10.58NCIITEE135 pKa = 4.03NEE137 pKa = 4.02VYY139 pKa = 10.59GIEE142 pKa = 4.34GSDD145 pKa = 3.72GEE147 pKa = 4.8GAGIYY152 pKa = 10.05CNNSSPRR159 pKa = 11.84IMDD162 pKa = 3.23CTISYY167 pKa = 10.58NIAAVSDD174 pKa = 3.53YY175 pKa = 10.93GYY177 pKa = 11.04AYY179 pKa = 10.46AHH181 pKa = 6.78GGGIYY186 pKa = 10.11SVQSSSTFTDD196 pKa = 3.43CTITHH201 pKa = 6.47NEE203 pKa = 3.62AGTFGGGISCWDD215 pKa = 3.49SSSSTFTDD223 pKa = 4.75CIISNNSALGRR234 pKa = 11.84GGGGIYY240 pKa = 10.28SSSTLMIEE248 pKa = 4.01RR249 pKa = 11.84CTVSDD254 pKa = 3.38NYY256 pKa = 10.87AYY258 pKa = 10.28QGGGIYY264 pKa = 9.99SYY266 pKa = 11.91GEE268 pKa = 4.0TTISDD273 pKa = 4.01SIIAGNSSDD282 pKa = 4.83DD283 pKa = 3.76DD284 pKa = 4.37GGGIFSYY291 pKa = 11.38GNILSLVNCTIFNNSGNVGGGVCFKK316 pKa = 10.6GDD318 pKa = 3.99CNATIANSIIADD330 pKa = 3.53NTGNCGGGIAVWGSPSGPGSFSITEE355 pKa = 4.07STISGNTASGCGGGFWFDD373 pKa = 3.75YY374 pKa = 11.05VPLATVTNCMIFRR387 pKa = 11.84NTALGGGGGGGNITGEE403 pKa = 4.37TSTMINSSTFFGNSSTEE420 pKa = 3.72VGGGIFYY427 pKa = 8.93RR428 pKa = 11.84TDD430 pKa = 3.32GDD432 pKa = 3.8NVSSFNVIKK441 pKa = 10.91NCILWDD447 pKa = 4.08DD448 pKa = 4.13SATDD452 pKa = 4.04GPEE455 pKa = 3.42IALVEE460 pKa = 4.37EE461 pKa = 4.64YY462 pKa = 10.96GDD464 pKa = 4.03PEE466 pKa = 4.62LSVEE470 pKa = 4.05MSYY473 pKa = 11.72SDD475 pKa = 3.63VKK477 pKa = 11.0GGQSDD482 pKa = 3.33VHH484 pKa = 8.22IIGGVLNWQAGNIEE498 pKa = 4.64SNPLFVDD505 pKa = 3.59PDD507 pKa = 3.94GDD509 pKa = 4.32DD510 pKa = 3.99NLPGTLDD517 pKa = 4.19DD518 pKa = 5.56DD519 pKa = 4.24FHH521 pKa = 6.42LTSDD525 pKa = 4.38SLCIDD530 pKa = 4.01AGTSQDD536 pKa = 4.01APSIDD541 pKa = 3.27IDD543 pKa = 4.19GDD545 pKa = 3.68PRR547 pKa = 11.84PQGAAIDD554 pKa = 3.96IGADD558 pKa = 3.52EE559 pKa = 5.38YY560 pKa = 10.98IDD562 pKa = 4.6PDD564 pKa = 3.66SDD566 pKa = 3.85GDD568 pKa = 4.14GIEE571 pKa = 4.06YY572 pKa = 10.55GIDD575 pKa = 3.06IDD577 pKa = 3.85PTVFSSEE584 pKa = 3.92FSDD587 pKa = 5.25AEE589 pKa = 4.09TTYY592 pKa = 11.22GIIIDD597 pKa = 4.48RR598 pKa = 11.84GDD600 pKa = 3.45QVVTVIDD607 pKa = 4.18LPDD610 pKa = 4.37PNGVRR615 pKa = 11.84IAASGGDD622 pKa = 3.65TPASVSACGGSAIFTLNDD640 pKa = 3.08GDD642 pKa = 5.12VITFTCGSVKK652 pKa = 10.04IEE654 pKa = 4.34VISGPVKK661 pKa = 10.24ISFVADD667 pKa = 3.89DD668 pKa = 4.15GTTATTTLDD677 pKa = 3.26AGNILTFEE685 pKa = 4.7PEE687 pKa = 4.15TLIITAPDD695 pKa = 3.64TNTEE699 pKa = 3.85NAVIVVDD706 pKa = 4.21GKK708 pKa = 11.06EE709 pKa = 3.81FAIEE713 pKa = 3.99PGEE716 pKa = 4.24SVEE719 pKa = 4.27LDD721 pKa = 2.98ITIVIDD727 pKa = 4.07GCDD730 pKa = 3.46TGVLDD735 pKa = 6.02SDD737 pKa = 5.11YY738 pKa = 10.45EE739 pKa = 4.14DD740 pKa = 3.71QSISEE745 pKa = 4.77WISQCAEE752 pKa = 3.49NAKK755 pKa = 10.02NHH757 pKa = 5.21GKK759 pKa = 10.04YY760 pKa = 10.01VSCVAKK766 pKa = 9.27LTKK769 pKa = 9.88KK770 pKa = 10.12LKK772 pKa = 9.47KK773 pKa = 10.43AGVITGKK780 pKa = 10.4EE781 pKa = 3.48KK782 pKa = 10.98GAIQSCAAQADD793 pKa = 3.61IHH795 pKa = 6.3
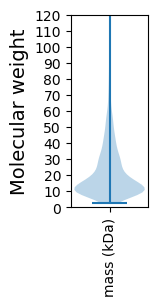
Molecular weight: 82.17 kDa
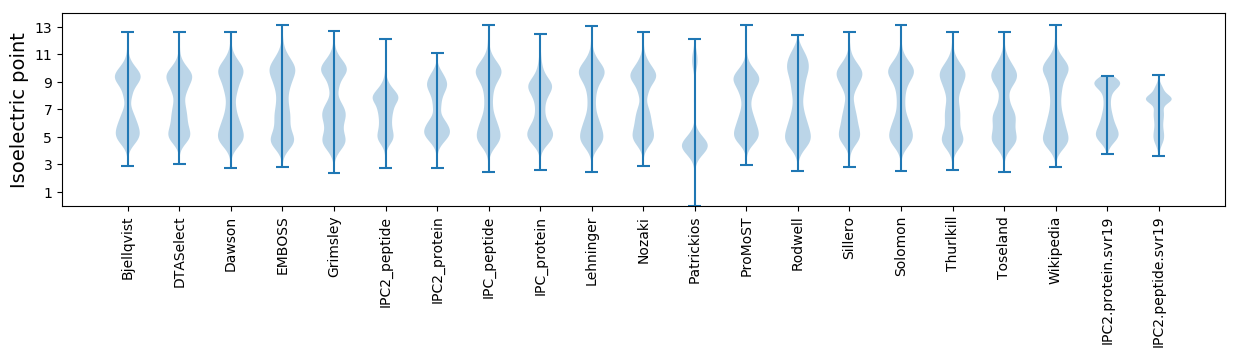
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2S8K265|A0A2S8K265_9DELT Uncharacterized protein (Fragment) OS=Desulfobacteraceae bacterium SEEP-SAG9 OX=2100161 GN=C6A37_13455 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.4RR3 pKa = 11.84TYY5 pKa = 9.91QPSRR9 pKa = 11.84IKK11 pKa = 10.5RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 6.4GFLKK20 pKa = 10.61RR21 pKa = 11.84MSTKK25 pKa = 9.49QGRR28 pKa = 11.84RR29 pKa = 11.84IINKK33 pKa = 8.92RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.48GRR39 pKa = 11.84KK40 pKa = 8.79RR41 pKa = 11.84LTVV44 pKa = 3.11
MM1 pKa = 7.35KK2 pKa = 9.4RR3 pKa = 11.84TYY5 pKa = 9.91QPSRR9 pKa = 11.84IKK11 pKa = 10.5RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 6.4GFLKK20 pKa = 10.61RR21 pKa = 11.84MSTKK25 pKa = 9.49QGRR28 pKa = 11.84RR29 pKa = 11.84IINKK33 pKa = 8.92RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.48GRR39 pKa = 11.84KK40 pKa = 8.79RR41 pKa = 11.84LTVV44 pKa = 3.11
Molecular weight: 5.37 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
550168 |
27 |
1467 |
209.9 |
23.54 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.528 ± 0.054 | 1.261 ± 0.023 |
5.392 ± 0.038 | 6.46 ± 0.066 |
4.61 ± 0.039 | 7.149 ± 0.062 |
2.095 ± 0.025 | 7.733 ± 0.051 |
6.663 ± 0.062 | 9.861 ± 0.058 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.604 ± 0.028 | 3.977 ± 0.033 |
4.29 ± 0.038 | 3.232 ± 0.029 |
5.363 ± 0.047 | 5.749 ± 0.036 |
5.048 ± 0.037 | 6.643 ± 0.04 |
1.148 ± 0.025 | 3.193 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
