
Human polyomavirus IPPyV
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Sepolyvirales; Polyomaviridae; Alphapolyomavirus; Human polyomavirus 9
Average proteome isoelectric point is 6.75
Get precalculated fractions of proteins
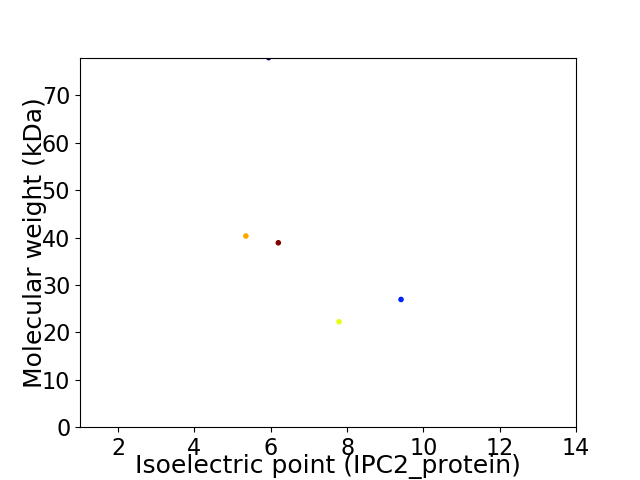
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F8K9Y5|F8K9Y5_9POLY Minor capsid protein OS=Human polyomavirus IPPyV OX=988774 GN=VP2 PE=3 SV=1
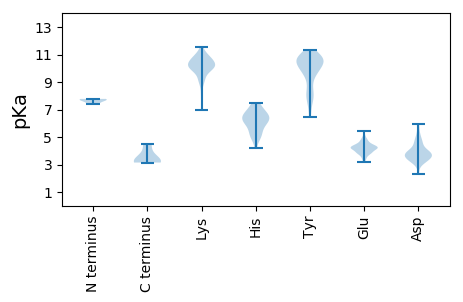
MM1 pKa = 7.58APQRR5 pKa = 11.84KK6 pKa = 8.37RR7 pKa = 11.84QEE9 pKa = 4.25CGACPVKK16 pKa = 10.01KK17 pKa = 8.84TCPTPAPVPKK27 pKa = 10.17LLVKK31 pKa = 10.79GGVEE35 pKa = 3.97VLEE38 pKa = 4.41VRR40 pKa = 11.84TGPDD44 pKa = 3.47AITQIEE50 pKa = 4.77AYY52 pKa = 10.29LNPRR56 pKa = 11.84MGNNNPTDD64 pKa = 3.56EE65 pKa = 4.81LYY67 pKa = 10.8GYY69 pKa = 10.12SADD72 pKa = 3.72INVASSKK79 pKa = 11.19ASDD82 pKa = 3.82NPNATTLPTYY92 pKa = 10.42SVAVIKK98 pKa = 10.8LPMLNEE104 pKa = 4.7DD105 pKa = 3.88MTCDD109 pKa = 3.43TLLMWEE115 pKa = 4.21AVSVKK120 pKa = 9.54TEE122 pKa = 3.61VMGISSLVNLHH133 pKa = 6.16QGGKK137 pKa = 9.56YY138 pKa = 9.87IYY140 pKa = 9.16GSSSGTIPVQGTTLHH155 pKa = 5.8MFSVGGEE162 pKa = 3.93PLEE165 pKa = 4.14LQGLVASSTTTYY177 pKa = 8.0PTDD180 pKa = 3.08MVTIKK185 pKa = 10.75NMKK188 pKa = 9.15PVNQALDD195 pKa = 3.93PNAKK199 pKa = 10.22ALLDD203 pKa = 3.78KK204 pKa = 10.92DD205 pKa = 3.32GKK207 pKa = 11.03YY208 pKa = 9.73PVEE211 pKa = 4.23VWSPDD216 pKa = 3.11PSKK219 pKa = 11.54NEE221 pKa = 3.36NTRR224 pKa = 11.84YY225 pKa = 10.01YY226 pKa = 11.35GSFTGGATTPPVMQFTNSVTTVLLDD251 pKa = 3.75EE252 pKa = 5.12NGVGPLCKK260 pKa = 9.66GDD262 pKa = 4.21KK263 pKa = 10.6LFLSAVDD270 pKa = 3.06IVGIHH275 pKa = 5.43TNYY278 pKa = 10.25SEE280 pKa = 4.2SQNWRR285 pKa = 11.84GLPRR289 pKa = 11.84YY290 pKa = 9.93FNVTLRR296 pKa = 11.84KK297 pKa = 9.25RR298 pKa = 11.84VVKK301 pKa = 10.6NPYY304 pKa = 9.06PVSSLLNSLFSGLMPQIQGQPMEE327 pKa = 4.48GTSGQVEE334 pKa = 4.43EE335 pKa = 4.02VRR337 pKa = 11.84IYY339 pKa = 10.71QGTEE343 pKa = 3.76GLPGDD348 pKa = 4.76PDD350 pKa = 3.8LDD352 pKa = 3.64RR353 pKa = 11.84YY354 pKa = 9.92VDD356 pKa = 4.37KK357 pKa = 10.98FCQNQTVPPRR367 pKa = 11.84SNDD370 pKa = 3.19QQ371 pKa = 3.14
MM1 pKa = 7.58APQRR5 pKa = 11.84KK6 pKa = 8.37RR7 pKa = 11.84QEE9 pKa = 4.25CGACPVKK16 pKa = 10.01KK17 pKa = 8.84TCPTPAPVPKK27 pKa = 10.17LLVKK31 pKa = 10.79GGVEE35 pKa = 3.97VLEE38 pKa = 4.41VRR40 pKa = 11.84TGPDD44 pKa = 3.47AITQIEE50 pKa = 4.77AYY52 pKa = 10.29LNPRR56 pKa = 11.84MGNNNPTDD64 pKa = 3.56EE65 pKa = 4.81LYY67 pKa = 10.8GYY69 pKa = 10.12SADD72 pKa = 3.72INVASSKK79 pKa = 11.19ASDD82 pKa = 3.82NPNATTLPTYY92 pKa = 10.42SVAVIKK98 pKa = 10.8LPMLNEE104 pKa = 4.7DD105 pKa = 3.88MTCDD109 pKa = 3.43TLLMWEE115 pKa = 4.21AVSVKK120 pKa = 9.54TEE122 pKa = 3.61VMGISSLVNLHH133 pKa = 6.16QGGKK137 pKa = 9.56YY138 pKa = 9.87IYY140 pKa = 9.16GSSSGTIPVQGTTLHH155 pKa = 5.8MFSVGGEE162 pKa = 3.93PLEE165 pKa = 4.14LQGLVASSTTTYY177 pKa = 8.0PTDD180 pKa = 3.08MVTIKK185 pKa = 10.75NMKK188 pKa = 9.15PVNQALDD195 pKa = 3.93PNAKK199 pKa = 10.22ALLDD203 pKa = 3.78KK204 pKa = 10.92DD205 pKa = 3.32GKK207 pKa = 11.03YY208 pKa = 9.73PVEE211 pKa = 4.23VWSPDD216 pKa = 3.11PSKK219 pKa = 11.54NEE221 pKa = 3.36NTRR224 pKa = 11.84YY225 pKa = 10.01YY226 pKa = 11.35GSFTGGATTPPVMQFTNSVTTVLLDD251 pKa = 3.75EE252 pKa = 5.12NGVGPLCKK260 pKa = 9.66GDD262 pKa = 4.21KK263 pKa = 10.6LFLSAVDD270 pKa = 3.06IVGIHH275 pKa = 5.43TNYY278 pKa = 10.25SEE280 pKa = 4.2SQNWRR285 pKa = 11.84GLPRR289 pKa = 11.84YY290 pKa = 9.93FNVTLRR296 pKa = 11.84KK297 pKa = 9.25RR298 pKa = 11.84VVKK301 pKa = 10.6NPYY304 pKa = 9.06PVSSLLNSLFSGLMPQIQGQPMEE327 pKa = 4.48GTSGQVEE334 pKa = 4.43EE335 pKa = 4.02VRR337 pKa = 11.84IYY339 pKa = 10.71QGTEE343 pKa = 3.76GLPGDD348 pKa = 4.76PDD350 pKa = 3.8LDD352 pKa = 3.64RR353 pKa = 11.84YY354 pKa = 9.92VDD356 pKa = 4.37KK357 pKa = 10.98FCQNQTVPPRR367 pKa = 11.84SNDD370 pKa = 3.19QQ371 pKa = 3.14
Molecular weight: 40.33 kDa
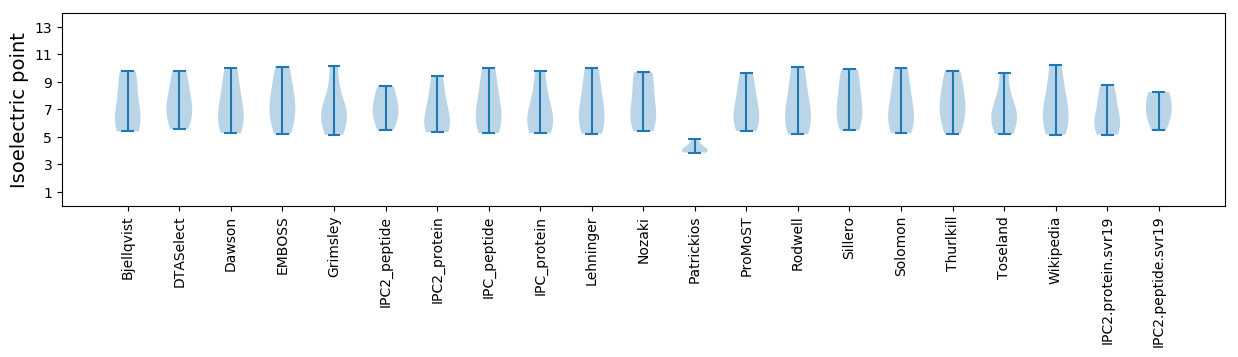
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F8K9Y6|F8K9Y6_9POLY Minor capsid protein OS=Human polyomavirus IPPyV OX=988774 GN=VP3 PE=3 SV=1
MM1 pKa = 7.7ALTPWFPQVDD11 pKa = 4.05YY12 pKa = 10.83LFPGLSSFSYY22 pKa = 10.36YY23 pKa = 10.81LNAALDD29 pKa = 3.48WGEE32 pKa = 4.4SLFHH36 pKa = 6.55AVGRR40 pKa = 11.84EE41 pKa = 3.21IWRR44 pKa = 11.84NIMRR48 pKa = 11.84QATQQIGYY56 pKa = 6.48TSRR59 pKa = 11.84ALAVRR64 pKa = 11.84GTNEE68 pKa = 4.05FQHH71 pKa = 6.21MLAQIAEE78 pKa = 4.27NARR81 pKa = 11.84WALTNGPIHH90 pKa = 7.04IYY92 pKa = 10.57SSVEE96 pKa = 3.49EE97 pKa = 4.47YY98 pKa = 11.07YY99 pKa = 10.71RR100 pKa = 11.84GLPSVNPIQLRR111 pKa = 11.84QQYY114 pKa = 9.51RR115 pKa = 11.84SRR117 pKa = 11.84GEE119 pKa = 3.83LPPTRR124 pKa = 11.84EE125 pKa = 3.5QFEE128 pKa = 4.27YY129 pKa = 10.75QEE131 pKa = 4.2QVRR134 pKa = 11.84LRR136 pKa = 11.84RR137 pKa = 11.84EE138 pKa = 3.78IGGSEE143 pKa = 4.21PRR145 pKa = 11.84SGHH148 pKa = 4.96YY149 pKa = 8.33VQHH152 pKa = 6.1YY153 pKa = 7.59AAPGGANQRR162 pKa = 11.84VSQDD166 pKa = 2.31WMLPLILGLYY176 pKa = 10.53GDD178 pKa = 4.72ITPTWEE184 pKa = 4.07KK185 pKa = 10.67EE186 pKa = 3.76LSKK189 pKa = 11.1LEE191 pKa = 4.0KK192 pKa = 10.47EE193 pKa = 4.58EE194 pKa = 3.92YY195 pKa = 10.71GPPKK199 pKa = 9.92KK200 pKa = 9.98KK201 pKa = 10.6ARR203 pKa = 11.84VRR205 pKa = 11.84SMSCKK210 pKa = 10.1KK211 pKa = 10.27NLSNTRR217 pKa = 11.84SRR219 pKa = 11.84SQTPCQRR226 pKa = 11.84RR227 pKa = 11.84GRR229 pKa = 11.84SSRR232 pKa = 11.84SS233 pKa = 3.16
MM1 pKa = 7.7ALTPWFPQVDD11 pKa = 4.05YY12 pKa = 10.83LFPGLSSFSYY22 pKa = 10.36YY23 pKa = 10.81LNAALDD29 pKa = 3.48WGEE32 pKa = 4.4SLFHH36 pKa = 6.55AVGRR40 pKa = 11.84EE41 pKa = 3.21IWRR44 pKa = 11.84NIMRR48 pKa = 11.84QATQQIGYY56 pKa = 6.48TSRR59 pKa = 11.84ALAVRR64 pKa = 11.84GTNEE68 pKa = 4.05FQHH71 pKa = 6.21MLAQIAEE78 pKa = 4.27NARR81 pKa = 11.84WALTNGPIHH90 pKa = 7.04IYY92 pKa = 10.57SSVEE96 pKa = 3.49EE97 pKa = 4.47YY98 pKa = 11.07YY99 pKa = 10.71RR100 pKa = 11.84GLPSVNPIQLRR111 pKa = 11.84QQYY114 pKa = 9.51RR115 pKa = 11.84SRR117 pKa = 11.84GEE119 pKa = 3.83LPPTRR124 pKa = 11.84EE125 pKa = 3.5QFEE128 pKa = 4.27YY129 pKa = 10.75QEE131 pKa = 4.2QVRR134 pKa = 11.84LRR136 pKa = 11.84RR137 pKa = 11.84EE138 pKa = 3.78IGGSEE143 pKa = 4.21PRR145 pKa = 11.84SGHH148 pKa = 4.96YY149 pKa = 8.33VQHH152 pKa = 6.1YY153 pKa = 7.59AAPGGANQRR162 pKa = 11.84VSQDD166 pKa = 2.31WMLPLILGLYY176 pKa = 10.53GDD178 pKa = 4.72ITPTWEE184 pKa = 4.07KK185 pKa = 10.67EE186 pKa = 3.76LSKK189 pKa = 11.1LEE191 pKa = 4.0KK192 pKa = 10.47EE193 pKa = 4.58EE194 pKa = 3.92YY195 pKa = 10.71GPPKK199 pKa = 9.92KK200 pKa = 9.98KK201 pKa = 10.6ARR203 pKa = 11.84VRR205 pKa = 11.84SMSCKK210 pKa = 10.1KK211 pKa = 10.27NLSNTRR217 pKa = 11.84SRR219 pKa = 11.84SQTPCQRR226 pKa = 11.84RR227 pKa = 11.84GRR229 pKa = 11.84SSRR232 pKa = 11.84SS233 pKa = 3.16
Molecular weight: 26.94 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1825 |
189 |
680 |
365.0 |
41.27 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.589 ± 1.142 | 2.137 ± 0.589 |
3.945 ± 0.763 | 6.849 ± 0.547 |
3.945 ± 0.726 | 6.63 ± 0.857 |
1.918 ± 0.471 | 4.712 ± 0.473 |
6.192 ± 1.393 | 10.466 ± 0.633 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.301 ± 0.284 | 4.822 ± 0.511 |
5.699 ± 0.745 | 5.096 ± 0.376 |
4.822 ± 1.179 | 7.671 ± 0.335 |
5.753 ± 0.707 | 5.589 ± 0.875 |
1.808 ± 0.346 | 4.055 ± 0.273 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
