
Helianthus annuus (Common sunflower)
Taxonomy: cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliopsida; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; campanulids; Asterales; Asteraceae; Asteroideae; Heliantheae alliance; Heliantheae; Helianthus
Average proteome isoelectric point is 6.94
Get precalculated fractions of proteins
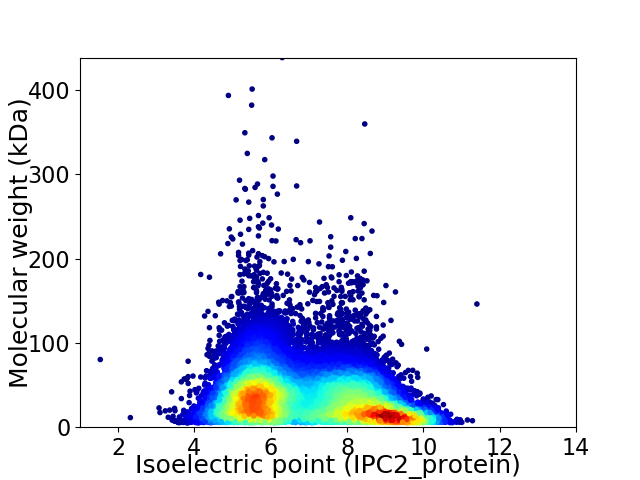
Virtual 2D-PAGE plot for 51240 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
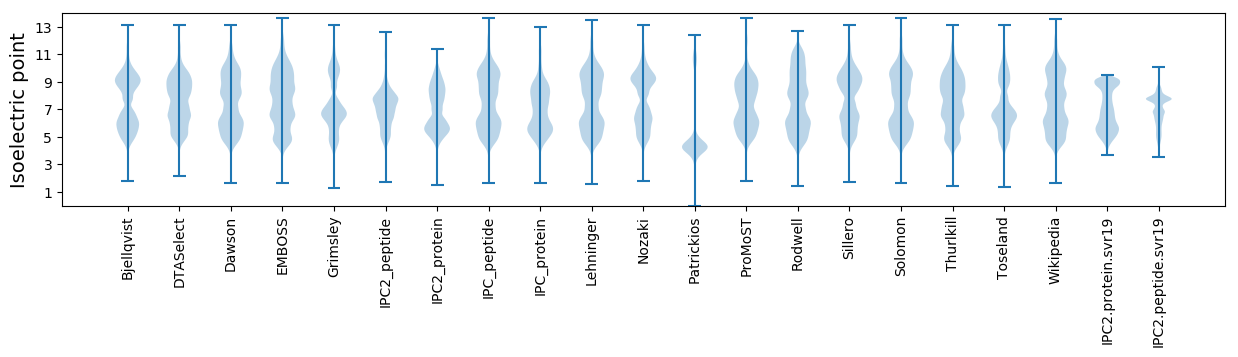
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A251TI58|A0A251TI58_HELAN Fatty acyl-CoA reductase OS=Helianthus annuus OX=4232 GN=HannXRQ_Chr10g0291761 PE=3 SV=1
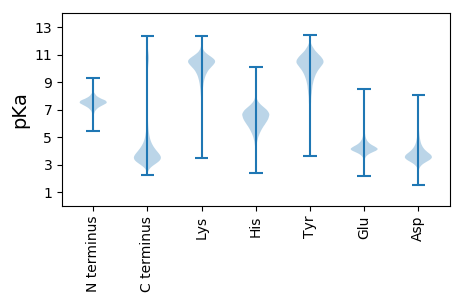
MM1 pKa = 7.62DD2 pKa = 4.15TSDD5 pKa = 4.22TGDD8 pKa = 3.3SDD10 pKa = 3.94TTGPLPMVSDD20 pKa = 5.27DD21 pKa = 3.61IVSSEE26 pKa = 4.59HH27 pKa = 6.11EE28 pKa = 4.49VHH30 pKa = 6.09TSDD33 pKa = 4.6YY34 pKa = 10.96TSTDD38 pKa = 3.38DD39 pKa = 6.61DD40 pKa = 4.43DD41 pKa = 4.69FQPFALPDD49 pKa = 3.92GADD52 pKa = 3.58EE53 pKa = 5.43PIVGDD58 pKa = 3.76PAEE61 pKa = 4.78DD62 pKa = 3.69LPLVVIPAPIPLASYY77 pKa = 8.76PAYY80 pKa = 10.12EE81 pKa = 4.38LMPGAEE87 pKa = 4.38ADD89 pKa = 3.49GDD91 pKa = 3.51IDD93 pKa = 4.81LFEE96 pKa = 5.54DD97 pKa = 4.15EE98 pKa = 4.77PLEE101 pKa = 5.43DD102 pKa = 4.29EE103 pKa = 4.87VPDD106 pKa = 4.43PALLPAGGLLMIAGAPVGDD125 pKa = 4.49SPVHH129 pKa = 6.02SPVPDD134 pKa = 3.35SCEE137 pKa = 4.14SVASAPSHH145 pKa = 5.38EE146 pKa = 4.47VSIQHH151 pKa = 6.07FVHH154 pKa = 7.63DD155 pKa = 4.98SDD157 pKa = 4.74PDD159 pKa = 3.67QASSAAPIPSLVFEE173 pKa = 4.97HH174 pKa = 7.42DD175 pKa = 4.07VIEE178 pKa = 5.08DD179 pKa = 3.7SDD181 pKa = 4.25PVFPPGFDD189 pKa = 3.49PDD191 pKa = 3.91RR192 pKa = 11.84DD193 pKa = 3.48IEE195 pKa = 4.54YY196 pKa = 10.37IPMDD200 pKa = 3.59HH201 pKa = 7.42HH202 pKa = 6.67MEE204 pKa = 4.64DD205 pKa = 3.71PVDD208 pKa = 3.92PVDD211 pKa = 5.27PIDD214 pKa = 5.58PIDD217 pKa = 4.22PDD219 pKa = 4.25FDD221 pKa = 4.85FDD223 pKa = 4.14MAFDD227 pKa = 4.31DD228 pKa = 4.94PEE230 pKa = 4.22PAVAPEE236 pKa = 3.55QAAAFDD242 pKa = 4.42PVHH245 pKa = 6.1EE246 pKa = 5.36HH247 pKa = 7.17DD248 pKa = 5.21LVHH251 pKa = 7.17ADD253 pKa = 3.63VPVDD257 pKa = 3.75PVLADD262 pKa = 3.88PPVGDD267 pKa = 4.84LPVDD271 pKa = 4.06DD272 pKa = 5.29VPLLADD278 pKa = 3.89DD279 pKa = 4.71HH280 pKa = 7.29VVDD283 pKa = 5.1PFVDD287 pKa = 4.05PPLIADD293 pKa = 3.73VPVDD297 pKa = 3.15HH298 pKa = 7.13HH299 pKa = 7.22VDD301 pKa = 3.21HH302 pKa = 7.23VDD304 pKa = 3.41PVIAPFDD311 pKa = 3.84LVPIEE316 pKa = 4.54PEE318 pKa = 3.48HH319 pKa = 7.75ALFAEE324 pKa = 4.81HH325 pKa = 7.04MDD327 pKa = 4.15PPDD330 pKa = 3.72EE331 pKa = 4.27EE332 pKa = 5.14AQHH335 pKa = 5.62GWIPADD341 pKa = 3.65EE342 pKa = 5.0DD343 pKa = 4.16VPPLPPHH350 pKa = 5.9HH351 pKa = 7.0TEE353 pKa = 3.48AHH355 pKa = 5.63HH356 pKa = 6.69TDD358 pKa = 3.86FSFQIPPFVPPAGPGEE374 pKa = 4.43GSSAHH379 pKa = 6.41PFGHH383 pKa = 6.65IPMPMPFMPQMTPVPSSVHH402 pKa = 5.12VAPFDD407 pKa = 3.78PTSTPLFWSSSSPMPPTDD425 pKa = 3.98PYY427 pKa = 11.15HH428 pKa = 6.48PFHH431 pKa = 6.83MGHH434 pKa = 6.67TIEE437 pKa = 4.87DD438 pKa = 3.55VLMSFVHH445 pKa = 5.27QHH447 pKa = 6.73DD448 pKa = 3.88SHH450 pKa = 6.0SQRR453 pKa = 11.84LQEE456 pKa = 4.53LEE458 pKa = 3.86RR459 pKa = 11.84AQLSFGPYY467 pKa = 10.1LGQTSSSFQPFRR479 pKa = 11.84SLPPDD484 pKa = 3.8FAARR488 pKa = 11.84LTTMEE493 pKa = 3.94QQIASVICTQQAMEE507 pKa = 4.69EE508 pKa = 4.25DD509 pKa = 3.87WLHH512 pKa = 6.42LRR514 pKa = 11.84RR515 pKa = 11.84LLYY518 pKa = 9.72THH520 pKa = 7.28FPPPPPPSAA529 pKa = 3.84
MM1 pKa = 7.62DD2 pKa = 4.15TSDD5 pKa = 4.22TGDD8 pKa = 3.3SDD10 pKa = 3.94TTGPLPMVSDD20 pKa = 5.27DD21 pKa = 3.61IVSSEE26 pKa = 4.59HH27 pKa = 6.11EE28 pKa = 4.49VHH30 pKa = 6.09TSDD33 pKa = 4.6YY34 pKa = 10.96TSTDD38 pKa = 3.38DD39 pKa = 6.61DD40 pKa = 4.43DD41 pKa = 4.69FQPFALPDD49 pKa = 3.92GADD52 pKa = 3.58EE53 pKa = 5.43PIVGDD58 pKa = 3.76PAEE61 pKa = 4.78DD62 pKa = 3.69LPLVVIPAPIPLASYY77 pKa = 8.76PAYY80 pKa = 10.12EE81 pKa = 4.38LMPGAEE87 pKa = 4.38ADD89 pKa = 3.49GDD91 pKa = 3.51IDD93 pKa = 4.81LFEE96 pKa = 5.54DD97 pKa = 4.15EE98 pKa = 4.77PLEE101 pKa = 5.43DD102 pKa = 4.29EE103 pKa = 4.87VPDD106 pKa = 4.43PALLPAGGLLMIAGAPVGDD125 pKa = 4.49SPVHH129 pKa = 6.02SPVPDD134 pKa = 3.35SCEE137 pKa = 4.14SVASAPSHH145 pKa = 5.38EE146 pKa = 4.47VSIQHH151 pKa = 6.07FVHH154 pKa = 7.63DD155 pKa = 4.98SDD157 pKa = 4.74PDD159 pKa = 3.67QASSAAPIPSLVFEE173 pKa = 4.97HH174 pKa = 7.42DD175 pKa = 4.07VIEE178 pKa = 5.08DD179 pKa = 3.7SDD181 pKa = 4.25PVFPPGFDD189 pKa = 3.49PDD191 pKa = 3.91RR192 pKa = 11.84DD193 pKa = 3.48IEE195 pKa = 4.54YY196 pKa = 10.37IPMDD200 pKa = 3.59HH201 pKa = 7.42HH202 pKa = 6.67MEE204 pKa = 4.64DD205 pKa = 3.71PVDD208 pKa = 3.92PVDD211 pKa = 5.27PIDD214 pKa = 5.58PIDD217 pKa = 4.22PDD219 pKa = 4.25FDD221 pKa = 4.85FDD223 pKa = 4.14MAFDD227 pKa = 4.31DD228 pKa = 4.94PEE230 pKa = 4.22PAVAPEE236 pKa = 3.55QAAAFDD242 pKa = 4.42PVHH245 pKa = 6.1EE246 pKa = 5.36HH247 pKa = 7.17DD248 pKa = 5.21LVHH251 pKa = 7.17ADD253 pKa = 3.63VPVDD257 pKa = 3.75PVLADD262 pKa = 3.88PPVGDD267 pKa = 4.84LPVDD271 pKa = 4.06DD272 pKa = 5.29VPLLADD278 pKa = 3.89DD279 pKa = 4.71HH280 pKa = 7.29VVDD283 pKa = 5.1PFVDD287 pKa = 4.05PPLIADD293 pKa = 3.73VPVDD297 pKa = 3.15HH298 pKa = 7.13HH299 pKa = 7.22VDD301 pKa = 3.21HH302 pKa = 7.23VDD304 pKa = 3.41PVIAPFDD311 pKa = 3.84LVPIEE316 pKa = 4.54PEE318 pKa = 3.48HH319 pKa = 7.75ALFAEE324 pKa = 4.81HH325 pKa = 7.04MDD327 pKa = 4.15PPDD330 pKa = 3.72EE331 pKa = 4.27EE332 pKa = 5.14AQHH335 pKa = 5.62GWIPADD341 pKa = 3.65EE342 pKa = 5.0DD343 pKa = 4.16VPPLPPHH350 pKa = 5.9HH351 pKa = 7.0TEE353 pKa = 3.48AHH355 pKa = 5.63HH356 pKa = 6.69TDD358 pKa = 3.86FSFQIPPFVPPAGPGEE374 pKa = 4.43GSSAHH379 pKa = 6.41PFGHH383 pKa = 6.65IPMPMPFMPQMTPVPSSVHH402 pKa = 5.12VAPFDD407 pKa = 3.78PTSTPLFWSSSSPMPPTDD425 pKa = 3.98PYY427 pKa = 11.15HH428 pKa = 6.48PFHH431 pKa = 6.83MGHH434 pKa = 6.67TIEE437 pKa = 4.87DD438 pKa = 3.55VLMSFVHH445 pKa = 5.27QHH447 pKa = 6.73DD448 pKa = 3.88SHH450 pKa = 6.0SQRR453 pKa = 11.84LQEE456 pKa = 4.53LEE458 pKa = 3.86RR459 pKa = 11.84AQLSFGPYY467 pKa = 10.1LGQTSSSFQPFRR479 pKa = 11.84SLPPDD484 pKa = 3.8FAARR488 pKa = 11.84LTTMEE493 pKa = 3.94QQIASVICTQQAMEE507 pKa = 4.69EE508 pKa = 4.25DD509 pKa = 3.87WLHH512 pKa = 6.42LRR514 pKa = 11.84RR515 pKa = 11.84LLYY518 pKa = 9.72THH520 pKa = 7.28FPPPPPPSAA529 pKa = 3.84
Molecular weight: 57.52 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A251SQ45|A0A251SQ45_HELAN Putative UDP-glucuronosyl/UDP-glucosyltransferase OS=Helianthus annuus OX=4232 GN=HannXRQ_Chr13g0393321 PE=3 SV=1
MM1 pKa = 7.16QLRR4 pKa = 11.84GKK6 pKa = 9.97LVLVGRR12 pKa = 11.84RR13 pKa = 11.84VPGARR18 pKa = 11.84VLATLKK24 pKa = 10.45LARR27 pKa = 11.84LRR29 pKa = 11.84LSQRR33 pKa = 11.84MKK35 pKa = 8.71RR36 pKa = 11.84WKK38 pKa = 10.32RR39 pKa = 11.84KK40 pKa = 8.54MPLPNLLGGRR50 pKa = 11.84GVGARR55 pKa = 11.84LQLLSLPRR63 pKa = 11.84LHH65 pKa = 6.93LWW67 pKa = 3.33
MM1 pKa = 7.16QLRR4 pKa = 11.84GKK6 pKa = 9.97LVLVGRR12 pKa = 11.84RR13 pKa = 11.84VPGARR18 pKa = 11.84VLATLKK24 pKa = 10.45LARR27 pKa = 11.84LRR29 pKa = 11.84LSQRR33 pKa = 11.84MKK35 pKa = 8.71RR36 pKa = 11.84WKK38 pKa = 10.32RR39 pKa = 11.84KK40 pKa = 8.54MPLPNLLGGRR50 pKa = 11.84GVGARR55 pKa = 11.84LQLLSLPRR63 pKa = 11.84LHH65 pKa = 6.93LWW67 pKa = 3.33
Molecular weight: 7.7 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
17848194 |
50 |
5399 |
348.3 |
39.05 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.115 ± 0.011 | 1.919 ± 0.006 |
5.358 ± 0.009 | 6.076 ± 0.011 |
4.397 ± 0.008 | 6.385 ± 0.011 |
2.513 ± 0.006 | 5.579 ± 0.01 |
6.258 ± 0.011 | 9.664 ± 0.013 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.568 ± 0.005 | 4.716 ± 0.008 |
4.785 ± 0.012 | 3.546 ± 0.007 |
5.115 ± 0.008 | 8.513 ± 0.013 |
5.319 ± 0.007 | 6.797 ± 0.01 |
1.338 ± 0.005 | 3.032 ± 0.006 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
