
Sophora japonica powdery mildew-associated partitivirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Partitiviridae; unclassified Partitiviridae
Average proteome isoelectric point is 6.98
Get precalculated fractions of proteins
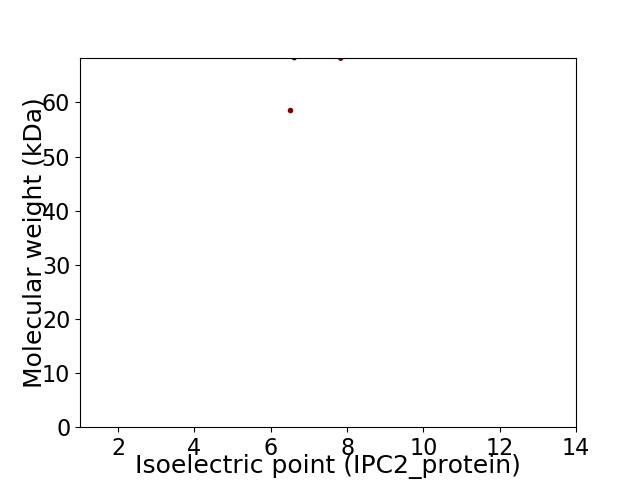
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1B3IZM6|A0A1B3IZM6_9VIRU Putative coat protein OS=Sophora japonica powdery mildew-associated partitivirus OX=1891704 PE=4 SV=1
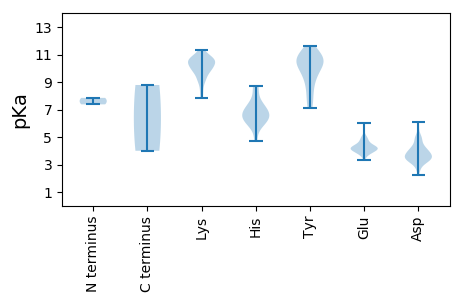
MM1 pKa = 7.38SGKK4 pKa = 7.88STKK7 pKa = 9.9KK8 pKa = 10.09SSSSKK13 pKa = 10.52SRR15 pKa = 11.84DD16 pKa = 3.26FRR18 pKa = 11.84KK19 pKa = 9.84EE20 pKa = 3.32AGRR23 pKa = 11.84PSLKK27 pKa = 10.24KK28 pKa = 10.65EE29 pKa = 3.85PVLSSGEE36 pKa = 4.18SSDD39 pKa = 4.25DD40 pKa = 3.62SSDD43 pKa = 4.09AEE45 pKa = 4.22IEE47 pKa = 4.1QALDD51 pKa = 3.33IPVKK55 pKa = 10.6DD56 pKa = 3.92DD57 pKa = 3.2SRR59 pKa = 11.84RR60 pKa = 11.84SKK62 pKa = 9.17STKK65 pKa = 9.05GKK67 pKa = 8.8QRR69 pKa = 11.84SVASRR74 pKa = 11.84EE75 pKa = 4.09KK76 pKa = 10.41KK77 pKa = 9.83QKK79 pKa = 10.07QKK81 pKa = 10.89PSVILADD88 pKa = 5.12DD89 pKa = 4.24DD90 pKa = 4.29TDD92 pKa = 5.7DD93 pKa = 4.85EE94 pKa = 6.02ANDD97 pKa = 3.85DD98 pKa = 4.02LAKK101 pKa = 10.6KK102 pKa = 10.21SEE104 pKa = 4.49SKK106 pKa = 10.44VAPHH110 pKa = 6.06MVLITGSTYY119 pKa = 10.49HH120 pKa = 6.55FGFPPVTHH128 pKa = 7.42PATSTFVPWCGNFFSVLFFMGEE150 pKa = 4.18SLCNSTLVQEE160 pKa = 4.87SIPDD164 pKa = 3.45YY165 pKa = 8.19FTPVLYY171 pKa = 9.58WYY173 pKa = 10.67GSMIFILQILRR184 pKa = 11.84ARR186 pKa = 11.84EE187 pKa = 3.54AAGAITRR194 pKa = 11.84DD195 pKa = 3.56EE196 pKa = 4.2RR197 pKa = 11.84TVLQMIIKK205 pKa = 8.33TYY207 pKa = 10.13PLEE210 pKa = 3.93QWTVLSPMIGFLQSLGSVKK229 pKa = 9.35PTDD232 pKa = 3.77PMYY235 pKa = 11.3SWICPAFPPFTGFTALHH252 pKa = 7.14SYY254 pKa = 10.65RR255 pKa = 11.84GMSTVPGIQRR265 pKa = 11.84MPPILAYY272 pKa = 10.28QKK274 pKa = 10.68FIFNFGAHH282 pKa = 4.74TTQTRR287 pKa = 11.84DD288 pKa = 3.0AFVYY292 pKa = 7.26PQPTITPQNQFVGLQGFGANDD313 pKa = 4.61DD314 pKa = 4.91DD315 pKa = 5.13FQCLTNMYY323 pKa = 9.44GWVSPPEE330 pKa = 4.22TGSLITVTDD339 pKa = 4.38LNLKK343 pKa = 9.67IRR345 pKa = 11.84TIRR348 pKa = 11.84RR349 pKa = 11.84WAVPDD354 pKa = 3.42VPNNLTVTNLKK365 pKa = 9.15TFMGLSDD372 pKa = 4.93DD373 pKa = 6.07LSLSWMFRR381 pKa = 11.84VLRR384 pKa = 11.84QMNSLSNFFPDD395 pKa = 3.34STNLANISPTTGLSSIADD413 pKa = 3.57VDD415 pKa = 3.46IGRR418 pKa = 11.84APARR422 pKa = 11.84IATDD426 pKa = 2.25SWYY429 pKa = 10.39RR430 pKa = 11.84GRR432 pKa = 11.84KK433 pKa = 9.23GITINILSPAGFDD446 pKa = 4.37DD447 pKa = 4.4SSQLRR452 pKa = 11.84LSAATCFNASLADD465 pKa = 3.88STHH468 pKa = 6.35SPLGRR473 pKa = 11.84QISSLQSGPYY483 pKa = 9.1FVNDD487 pKa = 3.78DD488 pKa = 4.24EE489 pKa = 4.76EE490 pKa = 4.69HH491 pKa = 6.69VSIPLIQYY499 pKa = 9.1EE500 pKa = 4.43SGARR504 pKa = 11.84TDD506 pKa = 3.32PVLRR510 pKa = 11.84FSEE513 pKa = 4.46IIEE516 pKa = 4.18SKK518 pKa = 10.89LFDD521 pKa = 3.91RR522 pKa = 11.84NGGRR526 pKa = 11.84KK527 pKa = 8.78
MM1 pKa = 7.38SGKK4 pKa = 7.88STKK7 pKa = 9.9KK8 pKa = 10.09SSSSKK13 pKa = 10.52SRR15 pKa = 11.84DD16 pKa = 3.26FRR18 pKa = 11.84KK19 pKa = 9.84EE20 pKa = 3.32AGRR23 pKa = 11.84PSLKK27 pKa = 10.24KK28 pKa = 10.65EE29 pKa = 3.85PVLSSGEE36 pKa = 4.18SSDD39 pKa = 4.25DD40 pKa = 3.62SSDD43 pKa = 4.09AEE45 pKa = 4.22IEE47 pKa = 4.1QALDD51 pKa = 3.33IPVKK55 pKa = 10.6DD56 pKa = 3.92DD57 pKa = 3.2SRR59 pKa = 11.84RR60 pKa = 11.84SKK62 pKa = 9.17STKK65 pKa = 9.05GKK67 pKa = 8.8QRR69 pKa = 11.84SVASRR74 pKa = 11.84EE75 pKa = 4.09KK76 pKa = 10.41KK77 pKa = 9.83QKK79 pKa = 10.07QKK81 pKa = 10.89PSVILADD88 pKa = 5.12DD89 pKa = 4.24DD90 pKa = 4.29TDD92 pKa = 5.7DD93 pKa = 4.85EE94 pKa = 6.02ANDD97 pKa = 3.85DD98 pKa = 4.02LAKK101 pKa = 10.6KK102 pKa = 10.21SEE104 pKa = 4.49SKK106 pKa = 10.44VAPHH110 pKa = 6.06MVLITGSTYY119 pKa = 10.49HH120 pKa = 6.55FGFPPVTHH128 pKa = 7.42PATSTFVPWCGNFFSVLFFMGEE150 pKa = 4.18SLCNSTLVQEE160 pKa = 4.87SIPDD164 pKa = 3.45YY165 pKa = 8.19FTPVLYY171 pKa = 9.58WYY173 pKa = 10.67GSMIFILQILRR184 pKa = 11.84ARR186 pKa = 11.84EE187 pKa = 3.54AAGAITRR194 pKa = 11.84DD195 pKa = 3.56EE196 pKa = 4.2RR197 pKa = 11.84TVLQMIIKK205 pKa = 8.33TYY207 pKa = 10.13PLEE210 pKa = 3.93QWTVLSPMIGFLQSLGSVKK229 pKa = 9.35PTDD232 pKa = 3.77PMYY235 pKa = 11.3SWICPAFPPFTGFTALHH252 pKa = 7.14SYY254 pKa = 10.65RR255 pKa = 11.84GMSTVPGIQRR265 pKa = 11.84MPPILAYY272 pKa = 10.28QKK274 pKa = 10.68FIFNFGAHH282 pKa = 4.74TTQTRR287 pKa = 11.84DD288 pKa = 3.0AFVYY292 pKa = 7.26PQPTITPQNQFVGLQGFGANDD313 pKa = 4.61DD314 pKa = 4.91DD315 pKa = 5.13FQCLTNMYY323 pKa = 9.44GWVSPPEE330 pKa = 4.22TGSLITVTDD339 pKa = 4.38LNLKK343 pKa = 9.67IRR345 pKa = 11.84TIRR348 pKa = 11.84RR349 pKa = 11.84WAVPDD354 pKa = 3.42VPNNLTVTNLKK365 pKa = 9.15TFMGLSDD372 pKa = 4.93DD373 pKa = 6.07LSLSWMFRR381 pKa = 11.84VLRR384 pKa = 11.84QMNSLSNFFPDD395 pKa = 3.34STNLANISPTTGLSSIADD413 pKa = 3.57VDD415 pKa = 3.46IGRR418 pKa = 11.84APARR422 pKa = 11.84IATDD426 pKa = 2.25SWYY429 pKa = 10.39RR430 pKa = 11.84GRR432 pKa = 11.84KK433 pKa = 9.23GITINILSPAGFDD446 pKa = 4.37DD447 pKa = 4.4SSQLRR452 pKa = 11.84LSAATCFNASLADD465 pKa = 3.88STHH468 pKa = 6.35SPLGRR473 pKa = 11.84QISSLQSGPYY483 pKa = 9.1FVNDD487 pKa = 3.78DD488 pKa = 4.24EE489 pKa = 4.76EE490 pKa = 4.69HH491 pKa = 6.69VSIPLIQYY499 pKa = 9.1EE500 pKa = 4.43SGARR504 pKa = 11.84TDD506 pKa = 3.32PVLRR510 pKa = 11.84FSEE513 pKa = 4.46IIEE516 pKa = 4.18SKK518 pKa = 10.89LFDD521 pKa = 3.91RR522 pKa = 11.84NGGRR526 pKa = 11.84KK527 pKa = 8.78
Molecular weight: 58.5 kDa
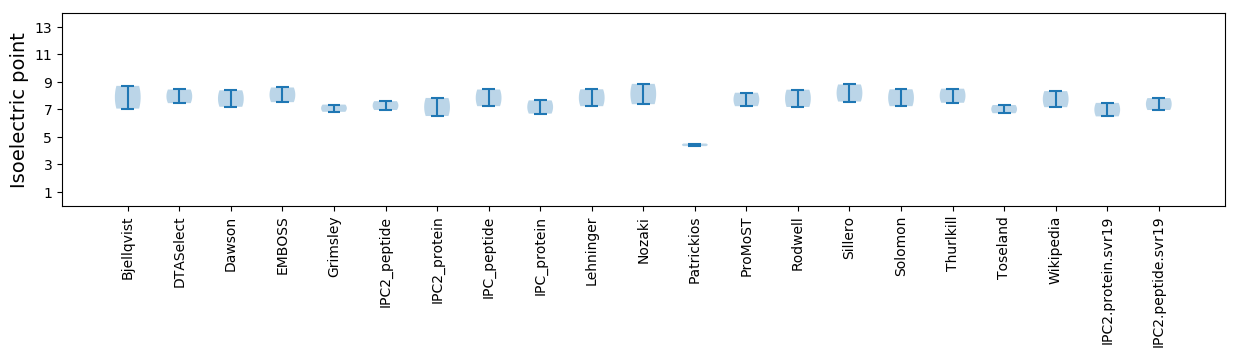
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1B3IZM6|A0A1B3IZM6_9VIRU Putative coat protein OS=Sophora japonica powdery mildew-associated partitivirus OX=1891704 PE=4 SV=1
MM1 pKa = 7.85ASTNLLRR8 pKa = 11.84LGKK11 pKa = 9.84IPRR14 pKa = 11.84SKK16 pKa = 10.5KK17 pKa = 10.19HH18 pKa = 6.25NDD20 pKa = 3.24PLSRR24 pKa = 11.84SLRR27 pKa = 11.84INRR30 pKa = 11.84IRR32 pKa = 11.84QGIIKK37 pKa = 9.97RR38 pKa = 11.84AIYY41 pKa = 9.36KK42 pKa = 9.74ICPINLARR50 pKa = 11.84QVIFGFKK57 pKa = 9.87RR58 pKa = 11.84SEE60 pKa = 4.15GSDD63 pKa = 3.3DD64 pKa = 3.61VAEE67 pKa = 4.17TDD69 pKa = 4.05FLRR72 pKa = 11.84SDD74 pKa = 3.5VPYY77 pKa = 10.93FDD79 pKa = 4.25MKK81 pKa = 10.94RR82 pKa = 11.84DD83 pKa = 3.66FHH85 pKa = 6.41YY86 pKa = 10.99LRR88 pKa = 11.84ALRR91 pKa = 11.84VCEE94 pKa = 3.73RR95 pKa = 11.84LFRR98 pKa = 11.84PSRR101 pKa = 11.84TLHH104 pKa = 6.58PIAFPDD110 pKa = 3.46LRR112 pKa = 11.84FYY114 pKa = 10.48PWSLSVSAEE123 pKa = 3.9APFSVEE129 pKa = 4.12KK130 pKa = 10.64KK131 pKa = 9.59RR132 pKa = 11.84STLIRR137 pKa = 11.84SRR139 pKa = 11.84QSDD142 pKa = 3.97GEE144 pKa = 4.22DD145 pKa = 3.19LDD147 pKa = 4.85GRR149 pKa = 11.84LTFHH153 pKa = 6.8NLYY156 pKa = 10.95NEE158 pKa = 3.79IFEE161 pKa = 4.97LNRR164 pKa = 11.84NLIHH168 pKa = 6.89QIKK171 pKa = 10.33EE172 pKa = 3.78GDD174 pKa = 3.21KK175 pKa = 11.3SFWNKK180 pKa = 10.72DD181 pKa = 3.45GTPRR185 pKa = 11.84PYY187 pKa = 10.04WYY189 pKa = 7.9NTLHH193 pKa = 5.92TRR195 pKa = 11.84PHH197 pKa = 6.0LVKK200 pKa = 10.71SSEE203 pKa = 3.95PDD205 pKa = 2.76KK206 pKa = 11.12LRR208 pKa = 11.84AVFGVPKK215 pKa = 10.18LLLMAEE221 pKa = 4.35NMFIWSLQKK230 pKa = 10.61EE231 pKa = 4.39YY232 pKa = 11.0LNQKK236 pKa = 8.42IQSPMLWGFEE246 pKa = 4.18TFKK249 pKa = 11.3GGWLKK254 pKa = 10.15IWNRR258 pKa = 11.84MYY260 pKa = 10.96SKK262 pKa = 10.55KK263 pKa = 10.54CSTFLSADD271 pKa = 2.53WSGFDD276 pKa = 3.3RR277 pKa = 11.84FALFEE282 pKa = 4.35CVDD285 pKa = 4.78DD286 pKa = 3.73IHH288 pKa = 8.73RR289 pKa = 11.84MWRR292 pKa = 11.84NWFDD296 pKa = 3.41FSKK299 pKa = 10.96YY300 pKa = 9.8EE301 pKa = 3.87PTIAEE306 pKa = 4.26SGPPGIQLSYY316 pKa = 10.26PKK318 pKa = 10.63SKK320 pKa = 9.87TNPQKK325 pKa = 10.36IEE327 pKa = 4.07RR328 pKa = 11.84LWNWMCYY335 pKa = 8.52CSKK338 pKa = 9.36YY339 pKa = 9.54TPIKK343 pKa = 9.1GQSGQLYY350 pKa = 7.49QWQYY354 pKa = 10.61NGIASGYY361 pKa = 8.53QQTQLIGSFVNSIYY375 pKa = 10.44MLTCLSDD382 pKa = 3.4LGINIEE388 pKa = 4.17SDD390 pKa = 2.99NFQLFVQGDD399 pKa = 3.63DD400 pKa = 3.54SLTEE404 pKa = 3.99FSEE407 pKa = 5.06IIQKK411 pKa = 10.76DD412 pKa = 4.02DD413 pKa = 3.62LPKK416 pKa = 10.68FLTNLAKK423 pKa = 10.26VAKK426 pKa = 10.2SRR428 pKa = 11.84FNANLSVQKK437 pKa = 7.84TTAGEE442 pKa = 4.1SLNDD446 pKa = 3.76VEE448 pKa = 4.8VLSYY452 pKa = 11.59NNTYY456 pKa = 10.88GIAFRR461 pKa = 11.84DD462 pKa = 3.7EE463 pKa = 4.62AEE465 pKa = 4.33LLAHH469 pKa = 7.07LLYY472 pKa = 10.28PEE474 pKa = 5.03RR475 pKa = 11.84FQTLEE480 pKa = 4.07ATVSCCIGIAYY491 pKa = 10.22ASMGCSQYY499 pKa = 11.19VDD501 pKa = 3.99DD502 pKa = 4.95TCLDD506 pKa = 3.53AYY508 pKa = 11.36NFLTTQFKK516 pKa = 10.69VKK518 pKa = 9.77PDD520 pKa = 3.62SNFLQDD526 pKa = 3.16FFRR529 pKa = 11.84IRR531 pKa = 11.84GSPLTDD537 pKa = 3.93DD538 pKa = 4.0FHH540 pKa = 8.6NPRR543 pKa = 11.84FPTKK547 pKa = 10.47DD548 pKa = 2.68EE549 pKa = 4.58CFNQNYY555 pKa = 7.11EE556 pKa = 3.88VRR558 pKa = 11.84SRR560 pKa = 11.84SDD562 pKa = 3.24SEE564 pKa = 4.3KK565 pKa = 9.91QRR567 pKa = 11.84LWPSIPTGDD576 pKa = 3.79YY577 pKa = 10.1GFHH580 pKa = 7.6FINEE584 pKa = 4.0
MM1 pKa = 7.85ASTNLLRR8 pKa = 11.84LGKK11 pKa = 9.84IPRR14 pKa = 11.84SKK16 pKa = 10.5KK17 pKa = 10.19HH18 pKa = 6.25NDD20 pKa = 3.24PLSRR24 pKa = 11.84SLRR27 pKa = 11.84INRR30 pKa = 11.84IRR32 pKa = 11.84QGIIKK37 pKa = 9.97RR38 pKa = 11.84AIYY41 pKa = 9.36KK42 pKa = 9.74ICPINLARR50 pKa = 11.84QVIFGFKK57 pKa = 9.87RR58 pKa = 11.84SEE60 pKa = 4.15GSDD63 pKa = 3.3DD64 pKa = 3.61VAEE67 pKa = 4.17TDD69 pKa = 4.05FLRR72 pKa = 11.84SDD74 pKa = 3.5VPYY77 pKa = 10.93FDD79 pKa = 4.25MKK81 pKa = 10.94RR82 pKa = 11.84DD83 pKa = 3.66FHH85 pKa = 6.41YY86 pKa = 10.99LRR88 pKa = 11.84ALRR91 pKa = 11.84VCEE94 pKa = 3.73RR95 pKa = 11.84LFRR98 pKa = 11.84PSRR101 pKa = 11.84TLHH104 pKa = 6.58PIAFPDD110 pKa = 3.46LRR112 pKa = 11.84FYY114 pKa = 10.48PWSLSVSAEE123 pKa = 3.9APFSVEE129 pKa = 4.12KK130 pKa = 10.64KK131 pKa = 9.59RR132 pKa = 11.84STLIRR137 pKa = 11.84SRR139 pKa = 11.84QSDD142 pKa = 3.97GEE144 pKa = 4.22DD145 pKa = 3.19LDD147 pKa = 4.85GRR149 pKa = 11.84LTFHH153 pKa = 6.8NLYY156 pKa = 10.95NEE158 pKa = 3.79IFEE161 pKa = 4.97LNRR164 pKa = 11.84NLIHH168 pKa = 6.89QIKK171 pKa = 10.33EE172 pKa = 3.78GDD174 pKa = 3.21KK175 pKa = 11.3SFWNKK180 pKa = 10.72DD181 pKa = 3.45GTPRR185 pKa = 11.84PYY187 pKa = 10.04WYY189 pKa = 7.9NTLHH193 pKa = 5.92TRR195 pKa = 11.84PHH197 pKa = 6.0LVKK200 pKa = 10.71SSEE203 pKa = 3.95PDD205 pKa = 2.76KK206 pKa = 11.12LRR208 pKa = 11.84AVFGVPKK215 pKa = 10.18LLLMAEE221 pKa = 4.35NMFIWSLQKK230 pKa = 10.61EE231 pKa = 4.39YY232 pKa = 11.0LNQKK236 pKa = 8.42IQSPMLWGFEE246 pKa = 4.18TFKK249 pKa = 11.3GGWLKK254 pKa = 10.15IWNRR258 pKa = 11.84MYY260 pKa = 10.96SKK262 pKa = 10.55KK263 pKa = 10.54CSTFLSADD271 pKa = 2.53WSGFDD276 pKa = 3.3RR277 pKa = 11.84FALFEE282 pKa = 4.35CVDD285 pKa = 4.78DD286 pKa = 3.73IHH288 pKa = 8.73RR289 pKa = 11.84MWRR292 pKa = 11.84NWFDD296 pKa = 3.41FSKK299 pKa = 10.96YY300 pKa = 9.8EE301 pKa = 3.87PTIAEE306 pKa = 4.26SGPPGIQLSYY316 pKa = 10.26PKK318 pKa = 10.63SKK320 pKa = 9.87TNPQKK325 pKa = 10.36IEE327 pKa = 4.07RR328 pKa = 11.84LWNWMCYY335 pKa = 8.52CSKK338 pKa = 9.36YY339 pKa = 9.54TPIKK343 pKa = 9.1GQSGQLYY350 pKa = 7.49QWQYY354 pKa = 10.61NGIASGYY361 pKa = 8.53QQTQLIGSFVNSIYY375 pKa = 10.44MLTCLSDD382 pKa = 3.4LGINIEE388 pKa = 4.17SDD390 pKa = 2.99NFQLFVQGDD399 pKa = 3.63DD400 pKa = 3.54SLTEE404 pKa = 3.99FSEE407 pKa = 5.06IIQKK411 pKa = 10.76DD412 pKa = 4.02DD413 pKa = 3.62LPKK416 pKa = 10.68FLTNLAKK423 pKa = 10.26VAKK426 pKa = 10.2SRR428 pKa = 11.84FNANLSVQKK437 pKa = 7.84TTAGEE442 pKa = 4.1SLNDD446 pKa = 3.76VEE448 pKa = 4.8VLSYY452 pKa = 11.59NNTYY456 pKa = 10.88GIAFRR461 pKa = 11.84DD462 pKa = 3.7EE463 pKa = 4.62AEE465 pKa = 4.33LLAHH469 pKa = 7.07LLYY472 pKa = 10.28PEE474 pKa = 5.03RR475 pKa = 11.84FQTLEE480 pKa = 4.07ATVSCCIGIAYY491 pKa = 10.22ASMGCSQYY499 pKa = 11.19VDD501 pKa = 3.99DD502 pKa = 4.95TCLDD506 pKa = 3.53AYY508 pKa = 11.36NFLTTQFKK516 pKa = 10.69VKK518 pKa = 9.77PDD520 pKa = 3.62SNFLQDD526 pKa = 3.16FFRR529 pKa = 11.84IRR531 pKa = 11.84GSPLTDD537 pKa = 3.93DD538 pKa = 4.0FHH540 pKa = 8.6NPRR543 pKa = 11.84FPTKK547 pKa = 10.47DD548 pKa = 2.68EE549 pKa = 4.58CFNQNYY555 pKa = 7.11EE556 pKa = 3.88VRR558 pKa = 11.84SRR560 pKa = 11.84SDD562 pKa = 3.24SEE564 pKa = 4.3KK565 pKa = 9.91QRR567 pKa = 11.84LWPSIPTGDD576 pKa = 3.79YY577 pKa = 10.1GFHH580 pKa = 7.6FINEE584 pKa = 4.0
Molecular weight: 68.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1111 |
527 |
584 |
555.5 |
63.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.041 ± 0.594 | 1.53 ± 0.411 |
6.571 ± 0.184 | 4.41 ± 0.569 |
6.301 ± 0.429 | 5.491 ± 0.411 |
1.62 ± 0.206 | 6.031 ± 0.029 |
5.581 ± 0.457 | 8.821 ± 0.601 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.07 ± 0.28 | 4.59 ± 0.696 |
5.851 ± 0.692 | 4.32 ± 0.103 |
6.121 ± 0.436 | 9.991 ± 1.119 |
6.031 ± 0.967 | 4.23 ± 0.631 |
1.98 ± 0.326 | 3.42 ± 0.673 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
