
Human gut microviridae SH-CHD8
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.92
Get precalculated fractions of proteins
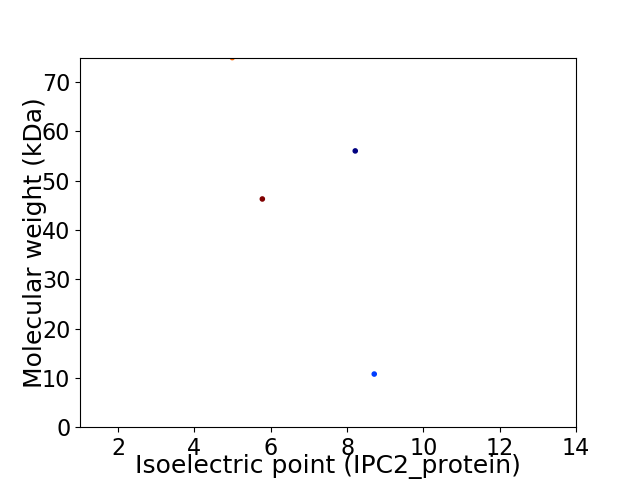
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
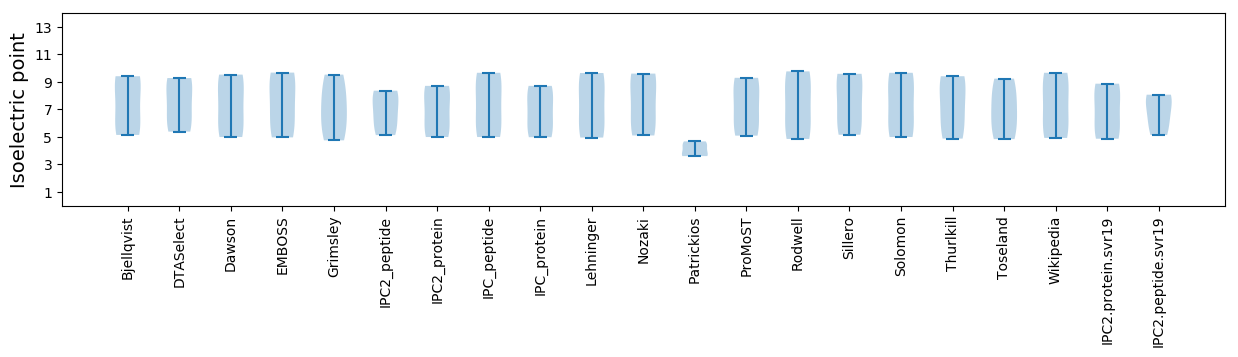
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1X9Q118|A0A1X9Q118_9VIRU Putative VP4 OS=Human gut microviridae SH-CHD8 OX=1986033 PE=4 SV=1
MM1 pKa = 7.56SNFSKK6 pKa = 10.49ISLGISTKK14 pKa = 10.22RR15 pKa = 11.84YY16 pKa = 5.98THH18 pKa = 7.45DD19 pKa = 3.13MSFDD23 pKa = 3.77NNTTLSFGVCQPLFCQILNDD43 pKa = 3.85KK44 pKa = 10.8DD45 pKa = 3.63KK46 pKa = 11.24LAGNIRR52 pKa = 11.84QLVRR56 pKa = 11.84LAPLPVPSFGRR67 pKa = 11.84MRR69 pKa = 11.84LVNKK73 pKa = 10.03LRR75 pKa = 11.84FVQMTDD81 pKa = 2.14ICPYY85 pKa = 9.3YY86 pKa = 10.59EE87 pKa = 4.58AMLSGAYY94 pKa = 10.58VNTSDD99 pKa = 5.82LNYY102 pKa = 10.48LPKK105 pKa = 10.55EE106 pKa = 4.05VLNDD110 pKa = 3.87SIKK113 pKa = 11.03VLTQMVLQYY122 pKa = 11.41SVFDD126 pKa = 3.91YY127 pKa = 10.77FKK129 pKa = 11.04VDD131 pKa = 3.61ASSSTPKK138 pKa = 10.47YY139 pKa = 10.02VYY141 pKa = 10.65QNVLTPSVLNEE152 pKa = 3.83ALSFYY157 pKa = 10.73HH158 pKa = 6.94LVVRR162 pKa = 11.84DD163 pKa = 3.66NTEE166 pKa = 3.14ISAAIPDD173 pKa = 4.37PKK175 pKa = 10.45PHH177 pKa = 6.6GGNVNTAISPDD188 pKa = 3.47SADD191 pKa = 3.66YY192 pKa = 11.0LISSDD197 pKa = 4.61SALICCRR204 pKa = 11.84LTEE207 pKa = 4.12RR208 pKa = 11.84GKK210 pKa = 10.39RR211 pKa = 11.84LRR213 pKa = 11.84NVLVGLGYY221 pKa = 11.04ALDD224 pKa = 3.99VSCTDD229 pKa = 3.55SVSLLPLFAYY239 pKa = 9.33FKK241 pKa = 10.81AWYY244 pKa = 9.5DD245 pKa = 3.23SYY247 pKa = 11.81APSRR251 pKa = 11.84EE252 pKa = 4.32TQWNQTNAFVLIDD265 pKa = 3.62ACFQNYY271 pKa = 7.33LTTLSGFTTANGSTQKK287 pKa = 9.86VQKK290 pKa = 10.51AVFGFFEE297 pKa = 4.81DD298 pKa = 5.47LANTWYY304 pKa = 10.23VAKK307 pKa = 10.25DD308 pKa = 3.79DD309 pKa = 4.05YY310 pKa = 11.88VSAHH314 pKa = 7.05RR315 pKa = 11.84LNAVSSNTDD324 pKa = 2.63SFGDD328 pKa = 3.72VQNKK332 pKa = 9.26IPSDD336 pKa = 3.83NNLTTLQGVKK346 pKa = 10.82YY347 pKa = 9.45NGNGNVPEE355 pKa = 4.19ATVVTDD361 pKa = 4.55FALQSLMRR369 pKa = 11.84ISKK372 pKa = 9.54FINKK376 pKa = 8.37DD377 pKa = 3.26TVIGKK382 pKa = 9.29KK383 pKa = 10.25VSTWLKK389 pKa = 8.83AHH391 pKa = 6.87FGADD395 pKa = 3.22VASSFFKK402 pKa = 10.89DD403 pKa = 3.83SVSLPDD409 pKa = 3.68VVVSCNINDD418 pKa = 3.84VFSTADD424 pKa = 3.07TSTSDD429 pKa = 3.86GSGEE433 pKa = 3.96QLGSYY438 pKa = 9.08AGKK441 pKa = 10.7GLGFGDD447 pKa = 4.03GSFSFTAPTFGYY459 pKa = 9.49FVVVSCLVPDD469 pKa = 3.83SKK471 pKa = 11.37YY472 pKa = 10.64FQGVDD477 pKa = 3.35PSLFGLDD484 pKa = 4.59RR485 pKa = 11.84FTLPNADD492 pKa = 4.43FDD494 pKa = 4.24ALSYY498 pKa = 10.79EE499 pKa = 3.92ITPCSFLCAHH509 pKa = 6.82NDD511 pKa = 3.46MYY513 pKa = 11.76VSGQKK518 pKa = 8.58STSDD522 pKa = 3.03KK523 pKa = 11.23GFGFVPRR530 pKa = 11.84YY531 pKa = 10.24SGFKK535 pKa = 8.1TRR537 pKa = 11.84RR538 pKa = 11.84NIVNGDD544 pKa = 3.15MSRR547 pKa = 11.84RR548 pKa = 11.84GTIASYY554 pKa = 10.88SPYY557 pKa = 10.58YY558 pKa = 9.88LDD560 pKa = 6.16KK561 pKa = 10.92ILTEE565 pKa = 3.71NSFRR569 pKa = 11.84SEE571 pKa = 3.84KK572 pKa = 10.63NADD575 pKa = 3.01GSYY578 pKa = 9.74TFVAVNNPLPLASEE592 pKa = 4.38VWRR595 pKa = 11.84FPTRR599 pKa = 11.84YY600 pKa = 9.26SWLGNFNRR608 pKa = 11.84IFYY611 pKa = 10.15NSGSAVFDD619 pKa = 4.3DD620 pKa = 4.76ASSSDD625 pKa = 3.62EE626 pKa = 4.23VNDD629 pKa = 3.76FTITPIDD636 pKa = 3.92DD637 pKa = 3.66NFIVQSVISMKK648 pKa = 9.61LTNALKK654 pKa = 9.86PIAMSYY660 pKa = 10.84DD661 pKa = 3.57VYY663 pKa = 11.58DD664 pKa = 5.13DD665 pKa = 5.67DD666 pKa = 5.59VDD668 pKa = 4.0NSSKK672 pKa = 10.09TITQEE677 pKa = 3.51
MM1 pKa = 7.56SNFSKK6 pKa = 10.49ISLGISTKK14 pKa = 10.22RR15 pKa = 11.84YY16 pKa = 5.98THH18 pKa = 7.45DD19 pKa = 3.13MSFDD23 pKa = 3.77NNTTLSFGVCQPLFCQILNDD43 pKa = 3.85KK44 pKa = 10.8DD45 pKa = 3.63KK46 pKa = 11.24LAGNIRR52 pKa = 11.84QLVRR56 pKa = 11.84LAPLPVPSFGRR67 pKa = 11.84MRR69 pKa = 11.84LVNKK73 pKa = 10.03LRR75 pKa = 11.84FVQMTDD81 pKa = 2.14ICPYY85 pKa = 9.3YY86 pKa = 10.59EE87 pKa = 4.58AMLSGAYY94 pKa = 10.58VNTSDD99 pKa = 5.82LNYY102 pKa = 10.48LPKK105 pKa = 10.55EE106 pKa = 4.05VLNDD110 pKa = 3.87SIKK113 pKa = 11.03VLTQMVLQYY122 pKa = 11.41SVFDD126 pKa = 3.91YY127 pKa = 10.77FKK129 pKa = 11.04VDD131 pKa = 3.61ASSSTPKK138 pKa = 10.47YY139 pKa = 10.02VYY141 pKa = 10.65QNVLTPSVLNEE152 pKa = 3.83ALSFYY157 pKa = 10.73HH158 pKa = 6.94LVVRR162 pKa = 11.84DD163 pKa = 3.66NTEE166 pKa = 3.14ISAAIPDD173 pKa = 4.37PKK175 pKa = 10.45PHH177 pKa = 6.6GGNVNTAISPDD188 pKa = 3.47SADD191 pKa = 3.66YY192 pKa = 11.0LISSDD197 pKa = 4.61SALICCRR204 pKa = 11.84LTEE207 pKa = 4.12RR208 pKa = 11.84GKK210 pKa = 10.39RR211 pKa = 11.84LRR213 pKa = 11.84NVLVGLGYY221 pKa = 11.04ALDD224 pKa = 3.99VSCTDD229 pKa = 3.55SVSLLPLFAYY239 pKa = 9.33FKK241 pKa = 10.81AWYY244 pKa = 9.5DD245 pKa = 3.23SYY247 pKa = 11.81APSRR251 pKa = 11.84EE252 pKa = 4.32TQWNQTNAFVLIDD265 pKa = 3.62ACFQNYY271 pKa = 7.33LTTLSGFTTANGSTQKK287 pKa = 9.86VQKK290 pKa = 10.51AVFGFFEE297 pKa = 4.81DD298 pKa = 5.47LANTWYY304 pKa = 10.23VAKK307 pKa = 10.25DD308 pKa = 3.79DD309 pKa = 4.05YY310 pKa = 11.88VSAHH314 pKa = 7.05RR315 pKa = 11.84LNAVSSNTDD324 pKa = 2.63SFGDD328 pKa = 3.72VQNKK332 pKa = 9.26IPSDD336 pKa = 3.83NNLTTLQGVKK346 pKa = 10.82YY347 pKa = 9.45NGNGNVPEE355 pKa = 4.19ATVVTDD361 pKa = 4.55FALQSLMRR369 pKa = 11.84ISKK372 pKa = 9.54FINKK376 pKa = 8.37DD377 pKa = 3.26TVIGKK382 pKa = 9.29KK383 pKa = 10.25VSTWLKK389 pKa = 8.83AHH391 pKa = 6.87FGADD395 pKa = 3.22VASSFFKK402 pKa = 10.89DD403 pKa = 3.83SVSLPDD409 pKa = 3.68VVVSCNINDD418 pKa = 3.84VFSTADD424 pKa = 3.07TSTSDD429 pKa = 3.86GSGEE433 pKa = 3.96QLGSYY438 pKa = 9.08AGKK441 pKa = 10.7GLGFGDD447 pKa = 4.03GSFSFTAPTFGYY459 pKa = 9.49FVVVSCLVPDD469 pKa = 3.83SKK471 pKa = 11.37YY472 pKa = 10.64FQGVDD477 pKa = 3.35PSLFGLDD484 pKa = 4.59RR485 pKa = 11.84FTLPNADD492 pKa = 4.43FDD494 pKa = 4.24ALSYY498 pKa = 10.79EE499 pKa = 3.92ITPCSFLCAHH509 pKa = 6.82NDD511 pKa = 3.46MYY513 pKa = 11.76VSGQKK518 pKa = 8.58STSDD522 pKa = 3.03KK523 pKa = 11.23GFGFVPRR530 pKa = 11.84YY531 pKa = 10.24SGFKK535 pKa = 8.1TRR537 pKa = 11.84RR538 pKa = 11.84NIVNGDD544 pKa = 3.15MSRR547 pKa = 11.84RR548 pKa = 11.84GTIASYY554 pKa = 10.88SPYY557 pKa = 10.58YY558 pKa = 9.88LDD560 pKa = 6.16KK561 pKa = 10.92ILTEE565 pKa = 3.71NSFRR569 pKa = 11.84SEE571 pKa = 3.84KK572 pKa = 10.63NADD575 pKa = 3.01GSYY578 pKa = 9.74TFVAVNNPLPLASEE592 pKa = 4.38VWRR595 pKa = 11.84FPTRR599 pKa = 11.84YY600 pKa = 9.26SWLGNFNRR608 pKa = 11.84IFYY611 pKa = 10.15NSGSAVFDD619 pKa = 4.3DD620 pKa = 4.76ASSSDD625 pKa = 3.62EE626 pKa = 4.23VNDD629 pKa = 3.76FTITPIDD636 pKa = 3.92DD637 pKa = 3.66NFIVQSVISMKK648 pKa = 9.61LTNALKK654 pKa = 9.86PIAMSYY660 pKa = 10.84DD661 pKa = 3.57VYY663 pKa = 11.58DD664 pKa = 5.13DD665 pKa = 5.67DD666 pKa = 5.59VDD668 pKa = 4.0NSSKK672 pKa = 10.09TITQEE677 pKa = 3.51
Molecular weight: 74.94 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1X9Q123|A0A1X9Q123_9VIRU Uncharacterized protein OS=Human gut microviridae SH-CHD8 OX=1986033 PE=4 SV=1
MM1 pKa = 7.8NYY3 pKa = 9.35PLPIVICLLILFMMNWSFSMGFCLNFLSKK32 pKa = 10.54FVMNKK37 pKa = 8.66EE38 pKa = 4.08VIKK41 pKa = 10.62IIIKK45 pKa = 9.18VVIYY49 pKa = 10.78ALGLIAAYY57 pKa = 10.46LGVTSLSSCTFSRR70 pKa = 11.84SSVAEE75 pKa = 3.92GRR77 pKa = 11.84ATIITTDD84 pKa = 3.26TTFISHH90 pKa = 6.38QGVLKK95 pKa = 10.18TKK97 pKa = 10.47
MM1 pKa = 7.8NYY3 pKa = 9.35PLPIVICLLILFMMNWSFSMGFCLNFLSKK32 pKa = 10.54FVMNKK37 pKa = 8.66EE38 pKa = 4.08VIKK41 pKa = 10.62IIIKK45 pKa = 9.18VVIYY49 pKa = 10.78ALGLIAAYY57 pKa = 10.46LGVTSLSSCTFSRR70 pKa = 11.84SSVAEE75 pKa = 3.92GRR77 pKa = 11.84ATIITTDD84 pKa = 3.26TTFISHH90 pKa = 6.38QGVLKK95 pKa = 10.18TKK97 pKa = 10.47
Molecular weight: 10.78 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1677 |
97 |
677 |
419.3 |
47.0 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.633 ± 1.96 | 1.968 ± 0.611 |
6.38 ± 0.79 | 3.637 ± 0.86 |
5.725 ± 0.919 | 5.605 ± 0.462 |
1.491 ± 0.488 | 4.651 ± 0.687 |
6.321 ± 0.737 | 9.064 ± 0.514 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.266 ± 0.362 | 6.5 ± 0.288 |
3.399 ± 0.505 | 3.936 ± 1.055 |
4.055 ± 0.473 | 9.362 ± 1.001 |
5.486 ± 0.837 | 6.44 ± 1.238 |
0.894 ± 0.022 | 5.188 ± 0.885 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
