
Tortoise microvirus 90
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.73
Get precalculated fractions of proteins
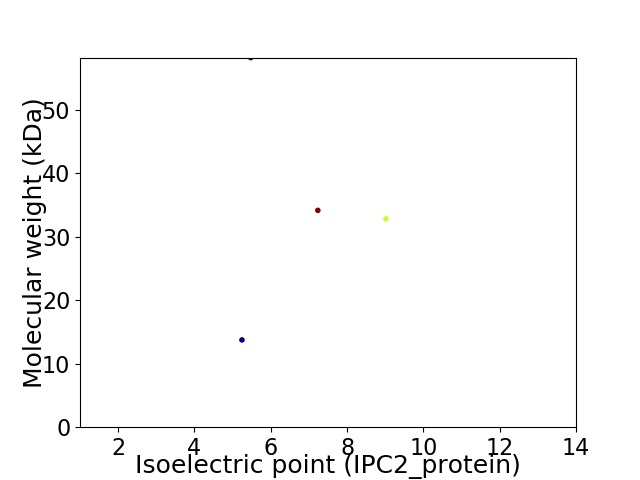
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W7V6|A0A4P8W7V6_9VIRU DNA pilot protein OS=Tortoise microvirus 90 OX=2583199 PE=4 SV=1
MM1 pKa = 7.15STTFALGEE9 pKa = 3.95QSTDD13 pKa = 3.32LEE15 pKa = 4.36KK16 pKa = 11.18SGGISQTVPDD26 pKa = 3.66QSLSLRR32 pKa = 11.84EE33 pKa = 3.88LVTRR37 pKa = 11.84QSQGLPIPMKK47 pKa = 10.6HH48 pKa = 6.69PIFSDD53 pKa = 3.67QEE55 pKa = 3.88IPDD58 pKa = 4.03LKK60 pKa = 10.97KK61 pKa = 10.73LDD63 pKa = 3.76LAEE66 pKa = 4.1IQEE69 pKa = 4.53LKK71 pKa = 10.43EE72 pKa = 3.99VAASQVNSLRR82 pKa = 11.84QKK84 pKa = 10.11LQQAEE89 pKa = 4.11LAEE92 pKa = 4.32RR93 pKa = 11.84QAVLAEE99 pKa = 4.14KK100 pKa = 10.42QKK102 pKa = 11.09NLDD105 pKa = 3.64KK106 pKa = 10.98AASAANQPPPAPPAQNSGVAATT128 pKa = 4.33
MM1 pKa = 7.15STTFALGEE9 pKa = 3.95QSTDD13 pKa = 3.32LEE15 pKa = 4.36KK16 pKa = 11.18SGGISQTVPDD26 pKa = 3.66QSLSLRR32 pKa = 11.84EE33 pKa = 3.88LVTRR37 pKa = 11.84QSQGLPIPMKK47 pKa = 10.6HH48 pKa = 6.69PIFSDD53 pKa = 3.67QEE55 pKa = 3.88IPDD58 pKa = 4.03LKK60 pKa = 10.97KK61 pKa = 10.73LDD63 pKa = 3.76LAEE66 pKa = 4.1IQEE69 pKa = 4.53LKK71 pKa = 10.43EE72 pKa = 3.99VAASQVNSLRR82 pKa = 11.84QKK84 pKa = 10.11LQQAEE89 pKa = 4.11LAEE92 pKa = 4.32RR93 pKa = 11.84QAVLAEE99 pKa = 4.14KK100 pKa = 10.42QKK102 pKa = 11.09NLDD105 pKa = 3.64KK106 pKa = 10.98AASAANQPPPAPPAQNSGVAATT128 pKa = 4.33
Molecular weight: 13.75 kDa
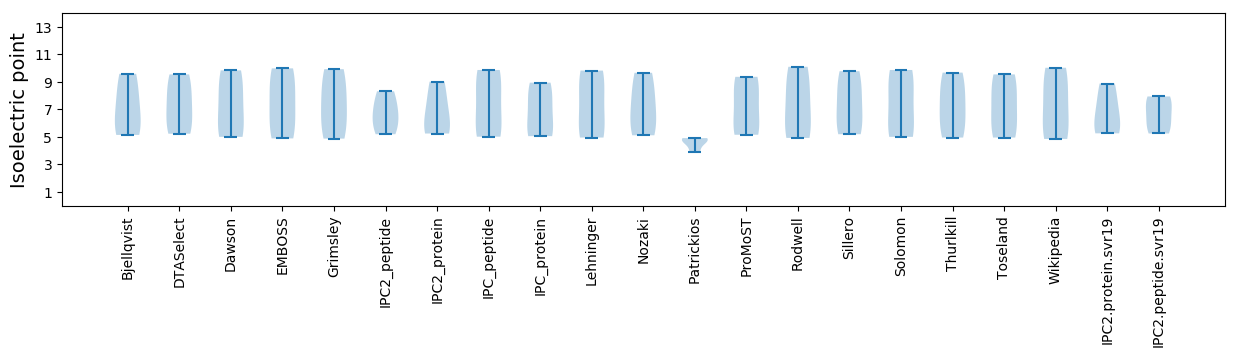
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8WAB2|A0A4P8WAB2_9VIRU Major capsid protein OS=Tortoise microvirus 90 OX=2583199 PE=3 SV=1
MM1 pKa = 7.54ACDD4 pKa = 4.43FPYY7 pKa = 10.48SIQNPTWPLRR17 pKa = 11.84SKK19 pKa = 9.76TKK21 pKa = 10.5FIPVPCGKK29 pKa = 10.33CPEE32 pKa = 4.24CCMRR36 pKa = 11.84RR37 pKa = 11.84SNNWQFRR44 pKa = 11.84LLQQEE49 pKa = 4.44KK50 pKa = 9.71ASSSASFVTLTYY62 pKa = 9.88NTDD65 pKa = 3.33HH66 pKa = 7.4VPMSKK71 pKa = 9.84RR72 pKa = 11.84GFMTLCKK79 pKa = 9.89TDD81 pKa = 3.79FQDD84 pKa = 3.21FMKK87 pKa = 10.52RR88 pKa = 11.84LRR90 pKa = 11.84YY91 pKa = 8.1YY92 pKa = 9.9HH93 pKa = 6.92PKK95 pKa = 9.28GAKK98 pKa = 8.65LRR100 pKa = 11.84YY101 pKa = 8.39YY102 pKa = 10.97AVGEE106 pKa = 4.24YY107 pKa = 10.57GSKK110 pKa = 8.61TFRR113 pKa = 11.84PHH115 pKa = 4.42FHH117 pKa = 7.28AIIFDD122 pKa = 4.28LDD124 pKa = 3.67RR125 pKa = 11.84EE126 pKa = 4.28QLPKK130 pKa = 10.7AWTLGDD136 pKa = 3.2IHH138 pKa = 8.54IGDD141 pKa = 3.62VSGASVAYY149 pKa = 4.99TTKK152 pKa = 10.71YY153 pKa = 6.42MHH155 pKa = 7.07KK156 pKa = 10.04GKK158 pKa = 9.65MIPVHH163 pKa = 6.83SNDD166 pKa = 3.15DD167 pKa = 4.19RR168 pKa = 11.84IPEE171 pKa = 4.02FSLMSKK177 pKa = 10.63KK178 pKa = 10.63LGANYY183 pKa = 7.76LTPQMVAYY191 pKa = 9.45HH192 pKa = 5.65QADD195 pKa = 3.59LTRR198 pKa = 11.84NYY200 pKa = 8.46VTLEE204 pKa = 4.13GGVKK208 pKa = 9.82VAMPRR213 pKa = 11.84YY214 pKa = 8.29YY215 pKa = 10.1RR216 pKa = 11.84EE217 pKa = 4.13KK218 pKa = 10.15IYY220 pKa = 11.01SEE222 pKa = 4.08EE223 pKa = 3.87QRR225 pKa = 11.84NLQNLHH231 pKa = 5.49NEE233 pKa = 4.04KK234 pKa = 10.15LAKK237 pKa = 9.96SALADD242 pKa = 3.54AEE244 pKa = 4.26EE245 pKa = 4.89SYY247 pKa = 10.97HH248 pKa = 6.71RR249 pKa = 11.84RR250 pKa = 11.84TGTSDD255 pKa = 3.43FEE257 pKa = 4.44RR258 pKa = 11.84ADD260 pKa = 3.6FEE262 pKa = 4.22ARR264 pKa = 11.84KK265 pKa = 10.03AKK267 pKa = 10.61LEE269 pKa = 3.92NFRR272 pKa = 11.84KK273 pKa = 9.78RR274 pKa = 11.84ARR276 pKa = 11.84DD277 pKa = 3.42GRR279 pKa = 11.84KK280 pKa = 9.9SII282 pKa = 4.29
MM1 pKa = 7.54ACDD4 pKa = 4.43FPYY7 pKa = 10.48SIQNPTWPLRR17 pKa = 11.84SKK19 pKa = 9.76TKK21 pKa = 10.5FIPVPCGKK29 pKa = 10.33CPEE32 pKa = 4.24CCMRR36 pKa = 11.84RR37 pKa = 11.84SNNWQFRR44 pKa = 11.84LLQQEE49 pKa = 4.44KK50 pKa = 9.71ASSSASFVTLTYY62 pKa = 9.88NTDD65 pKa = 3.33HH66 pKa = 7.4VPMSKK71 pKa = 9.84RR72 pKa = 11.84GFMTLCKK79 pKa = 9.89TDD81 pKa = 3.79FQDD84 pKa = 3.21FMKK87 pKa = 10.52RR88 pKa = 11.84LRR90 pKa = 11.84YY91 pKa = 8.1YY92 pKa = 9.9HH93 pKa = 6.92PKK95 pKa = 9.28GAKK98 pKa = 8.65LRR100 pKa = 11.84YY101 pKa = 8.39YY102 pKa = 10.97AVGEE106 pKa = 4.24YY107 pKa = 10.57GSKK110 pKa = 8.61TFRR113 pKa = 11.84PHH115 pKa = 4.42FHH117 pKa = 7.28AIIFDD122 pKa = 4.28LDD124 pKa = 3.67RR125 pKa = 11.84EE126 pKa = 4.28QLPKK130 pKa = 10.7AWTLGDD136 pKa = 3.2IHH138 pKa = 8.54IGDD141 pKa = 3.62VSGASVAYY149 pKa = 4.99TTKK152 pKa = 10.71YY153 pKa = 6.42MHH155 pKa = 7.07KK156 pKa = 10.04GKK158 pKa = 9.65MIPVHH163 pKa = 6.83SNDD166 pKa = 3.15DD167 pKa = 4.19RR168 pKa = 11.84IPEE171 pKa = 4.02FSLMSKK177 pKa = 10.63KK178 pKa = 10.63LGANYY183 pKa = 7.76LTPQMVAYY191 pKa = 9.45HH192 pKa = 5.65QADD195 pKa = 3.59LTRR198 pKa = 11.84NYY200 pKa = 8.46VTLEE204 pKa = 4.13GGVKK208 pKa = 9.82VAMPRR213 pKa = 11.84YY214 pKa = 8.29YY215 pKa = 10.1RR216 pKa = 11.84EE217 pKa = 4.13KK218 pKa = 10.15IYY220 pKa = 11.01SEE222 pKa = 4.08EE223 pKa = 3.87QRR225 pKa = 11.84NLQNLHH231 pKa = 5.49NEE233 pKa = 4.04KK234 pKa = 10.15LAKK237 pKa = 9.96SALADD242 pKa = 3.54AEE244 pKa = 4.26EE245 pKa = 4.89SYY247 pKa = 10.97HH248 pKa = 6.71RR249 pKa = 11.84RR250 pKa = 11.84TGTSDD255 pKa = 3.43FEE257 pKa = 4.44RR258 pKa = 11.84ADD260 pKa = 3.6FEE262 pKa = 4.22ARR264 pKa = 11.84KK265 pKa = 10.03AKK267 pKa = 10.61LEE269 pKa = 3.92NFRR272 pKa = 11.84KK273 pKa = 9.78RR274 pKa = 11.84ARR276 pKa = 11.84DD277 pKa = 3.42GRR279 pKa = 11.84KK280 pKa = 9.9SII282 pKa = 4.29
Molecular weight: 32.83 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1235 |
128 |
519 |
308.8 |
34.75 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.854 ± 1.069 | 0.567 ± 0.427 |
5.668 ± 0.24 | 5.911 ± 0.415 |
3.968 ± 1.076 | 6.316 ± 1.04 |
2.51 ± 0.816 | 5.263 ± 0.767 |
5.911 ± 0.82 | 8.259 ± 1.298 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.672 ± 0.277 | 5.02 ± 0.785 |
5.668 ± 0.709 | 5.425 ± 1.477 |
5.83 ± 0.899 | 7.935 ± 1.212 |
5.182 ± 0.36 | 4.858 ± 0.644 |
0.81 ± 0.272 | 4.372 ± 0.976 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
