
Bacteroides sp. CAG:598
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; environmental samples
Average proteome isoelectric point is 6.35
Get precalculated fractions of proteins
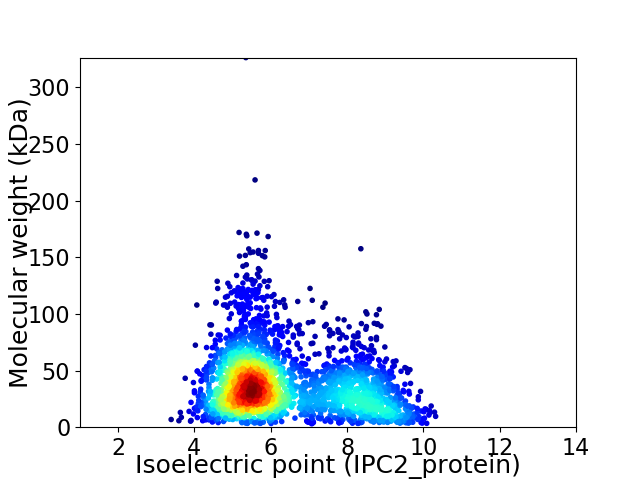
Virtual 2D-PAGE plot for 2776 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R5C4Q5|R5C4Q5_9BACE Uncharacterized protein OS=Bacteroides sp. CAG:598 OX=1262743 GN=BN727_00959 PE=4 SV=1
MM1 pKa = 7.27YY2 pKa = 10.33KK3 pKa = 9.12RR4 pKa = 11.84TFMQTLAAVAMMLLPAACADD24 pKa = 4.55DD25 pKa = 4.67YY26 pKa = 11.86DD27 pKa = 6.46DD28 pKa = 4.08SALWDD33 pKa = 3.52KK34 pKa = 11.66VNDD37 pKa = 3.69HH38 pKa = 6.3EE39 pKa = 4.58EE40 pKa = 3.74RR41 pKa = 11.84LAALEE46 pKa = 3.96KK47 pKa = 10.09WQEE50 pKa = 3.89QTNQSIAAMQEE61 pKa = 4.14LLNTTDD67 pKa = 3.57MITGVSAVSEE77 pKa = 4.46GGQTVGYY84 pKa = 8.23TITFLHH90 pKa = 6.93SDD92 pKa = 4.91PITIYY97 pKa = 10.82NGAKK101 pKa = 10.15GADD104 pKa = 3.95GEE106 pKa = 4.76DD107 pKa = 3.97GADD110 pKa = 3.73GADD113 pKa = 3.47GQTPQIGLAQAEE125 pKa = 4.35DD126 pKa = 4.52GNWYY130 pKa = 6.21WTLNGEE136 pKa = 4.98LLTDD140 pKa = 3.7AQGNPIRR147 pKa = 11.84ANGLDD152 pKa = 3.77GADD155 pKa = 3.88GADD158 pKa = 3.99GEE160 pKa = 5.04DD161 pKa = 4.26GATGPEE167 pKa = 4.57GPQGNDD173 pKa = 2.98GTSAPTPQILLGSSITSGTIKK194 pKa = 9.88TDD196 pKa = 3.0NGQKK200 pKa = 9.34DD201 pKa = 2.98TDD203 pKa = 3.15AWYY206 pKa = 10.77LSVDD210 pKa = 4.55NGATWYY216 pKa = 10.22RR217 pKa = 11.84VSGKK221 pKa = 10.42DD222 pKa = 3.43GTNGDD227 pKa = 3.38AWFSDD232 pKa = 3.9EE233 pKa = 4.48PKK235 pKa = 10.48KK236 pKa = 10.72EE237 pKa = 3.52GNYY240 pKa = 10.1YY241 pKa = 10.71VFTLTDD247 pKa = 3.06QSTFRR252 pKa = 11.84VAAYY256 pKa = 9.82QPFRR260 pKa = 11.84ILTEE264 pKa = 3.97AEE266 pKa = 3.91KK267 pKa = 11.06DD268 pKa = 3.39GTTTFTNATVTINGATTFYY287 pKa = 10.99LYY289 pKa = 10.19IDD291 pKa = 3.88NDD293 pKa = 3.05IKK295 pKa = 10.92YY296 pKa = 9.88QEE298 pKa = 4.06IVAQISSVDD307 pKa = 3.53AVLTRR312 pKa = 11.84ANVSDD317 pKa = 3.32WTVVKK322 pKa = 9.75GTEE325 pKa = 4.11QNTLTVTPGTQPYY338 pKa = 10.59ALLDD342 pKa = 3.69VSLLLTDD349 pKa = 4.52GSKK352 pKa = 9.23LTASRR357 pKa = 11.84LLEE360 pKa = 4.57DD361 pKa = 4.71PGYY364 pKa = 9.65TDD366 pKa = 5.66DD367 pKa = 4.22GQGNYY372 pKa = 7.72TVTSAEE378 pKa = 3.95GLKK381 pKa = 10.85NIAKK385 pKa = 9.94LVNEE389 pKa = 4.41EE390 pKa = 4.16GKK392 pKa = 9.01TDD394 pKa = 3.57INITLDD400 pKa = 3.19TDD402 pKa = 3.83LTLTGEE408 pKa = 3.99WTPIGTEE415 pKa = 3.94SQPYY419 pKa = 9.24TGTFDD424 pKa = 4.06GGNHH428 pKa = 7.26TITGLKK434 pKa = 9.16IDD436 pKa = 4.07QSGTDD441 pKa = 3.49NVGLIGRR448 pKa = 11.84LGSGGKK454 pKa = 9.09VQDD457 pKa = 3.64VTLTEE462 pKa = 4.22VNVTGGTYY470 pKa = 10.2VGGIAGQTDD479 pKa = 3.94GTVEE483 pKa = 3.93NCSVNGTVTGQNQTGGIVGRR503 pKa = 11.84NFSTISGCSAEE514 pKa = 4.25GTVTGNTNVGGISGLCVPNYY534 pKa = 8.06DD535 pKa = 3.82TGTGSLIGSTIEE547 pKa = 4.47GCHH550 pKa = 4.72STAAVSGISSVGGVVGNLGNGCSLMACYY578 pKa = 9.38STGNVTATITYY589 pKa = 8.56GHH591 pKa = 7.02ANVGGVVGINGQGTVTACYY610 pKa = 9.43HH611 pKa = 5.28ATGEE615 pKa = 4.39ITSSVGDD622 pKa = 4.01RR623 pKa = 11.84IGGIAGCNDD632 pKa = 2.93QGTFTACYY640 pKa = 8.88WEE642 pKa = 4.3NNQEE646 pKa = 3.97QGTGSGSTGGTAKK659 pKa = 10.75VEE661 pKa = 4.49GNVIWKK667 pKa = 8.03TAVDD671 pKa = 3.78AMNAALNKK679 pKa = 10.53SGWTYY684 pKa = 11.48RR685 pKa = 11.84LGGGNLPEE693 pKa = 4.67LVQNQQ698 pKa = 3.27
MM1 pKa = 7.27YY2 pKa = 10.33KK3 pKa = 9.12RR4 pKa = 11.84TFMQTLAAVAMMLLPAACADD24 pKa = 4.55DD25 pKa = 4.67YY26 pKa = 11.86DD27 pKa = 6.46DD28 pKa = 4.08SALWDD33 pKa = 3.52KK34 pKa = 11.66VNDD37 pKa = 3.69HH38 pKa = 6.3EE39 pKa = 4.58EE40 pKa = 3.74RR41 pKa = 11.84LAALEE46 pKa = 3.96KK47 pKa = 10.09WQEE50 pKa = 3.89QTNQSIAAMQEE61 pKa = 4.14LLNTTDD67 pKa = 3.57MITGVSAVSEE77 pKa = 4.46GGQTVGYY84 pKa = 8.23TITFLHH90 pKa = 6.93SDD92 pKa = 4.91PITIYY97 pKa = 10.82NGAKK101 pKa = 10.15GADD104 pKa = 3.95GEE106 pKa = 4.76DD107 pKa = 3.97GADD110 pKa = 3.73GADD113 pKa = 3.47GQTPQIGLAQAEE125 pKa = 4.35DD126 pKa = 4.52GNWYY130 pKa = 6.21WTLNGEE136 pKa = 4.98LLTDD140 pKa = 3.7AQGNPIRR147 pKa = 11.84ANGLDD152 pKa = 3.77GADD155 pKa = 3.88GADD158 pKa = 3.99GEE160 pKa = 5.04DD161 pKa = 4.26GATGPEE167 pKa = 4.57GPQGNDD173 pKa = 2.98GTSAPTPQILLGSSITSGTIKK194 pKa = 9.88TDD196 pKa = 3.0NGQKK200 pKa = 9.34DD201 pKa = 2.98TDD203 pKa = 3.15AWYY206 pKa = 10.77LSVDD210 pKa = 4.55NGATWYY216 pKa = 10.22RR217 pKa = 11.84VSGKK221 pKa = 10.42DD222 pKa = 3.43GTNGDD227 pKa = 3.38AWFSDD232 pKa = 3.9EE233 pKa = 4.48PKK235 pKa = 10.48KK236 pKa = 10.72EE237 pKa = 3.52GNYY240 pKa = 10.1YY241 pKa = 10.71VFTLTDD247 pKa = 3.06QSTFRR252 pKa = 11.84VAAYY256 pKa = 9.82QPFRR260 pKa = 11.84ILTEE264 pKa = 3.97AEE266 pKa = 3.91KK267 pKa = 11.06DD268 pKa = 3.39GTTTFTNATVTINGATTFYY287 pKa = 10.99LYY289 pKa = 10.19IDD291 pKa = 3.88NDD293 pKa = 3.05IKK295 pKa = 10.92YY296 pKa = 9.88QEE298 pKa = 4.06IVAQISSVDD307 pKa = 3.53AVLTRR312 pKa = 11.84ANVSDD317 pKa = 3.32WTVVKK322 pKa = 9.75GTEE325 pKa = 4.11QNTLTVTPGTQPYY338 pKa = 10.59ALLDD342 pKa = 3.69VSLLLTDD349 pKa = 4.52GSKK352 pKa = 9.23LTASRR357 pKa = 11.84LLEE360 pKa = 4.57DD361 pKa = 4.71PGYY364 pKa = 9.65TDD366 pKa = 5.66DD367 pKa = 4.22GQGNYY372 pKa = 7.72TVTSAEE378 pKa = 3.95GLKK381 pKa = 10.85NIAKK385 pKa = 9.94LVNEE389 pKa = 4.41EE390 pKa = 4.16GKK392 pKa = 9.01TDD394 pKa = 3.57INITLDD400 pKa = 3.19TDD402 pKa = 3.83LTLTGEE408 pKa = 3.99WTPIGTEE415 pKa = 3.94SQPYY419 pKa = 9.24TGTFDD424 pKa = 4.06GGNHH428 pKa = 7.26TITGLKK434 pKa = 9.16IDD436 pKa = 4.07QSGTDD441 pKa = 3.49NVGLIGRR448 pKa = 11.84LGSGGKK454 pKa = 9.09VQDD457 pKa = 3.64VTLTEE462 pKa = 4.22VNVTGGTYY470 pKa = 10.2VGGIAGQTDD479 pKa = 3.94GTVEE483 pKa = 3.93NCSVNGTVTGQNQTGGIVGRR503 pKa = 11.84NFSTISGCSAEE514 pKa = 4.25GTVTGNTNVGGISGLCVPNYY534 pKa = 8.06DD535 pKa = 3.82TGTGSLIGSTIEE547 pKa = 4.47GCHH550 pKa = 4.72STAAVSGISSVGGVVGNLGNGCSLMACYY578 pKa = 9.38STGNVTATITYY589 pKa = 8.56GHH591 pKa = 7.02ANVGGVVGINGQGTVTACYY610 pKa = 9.43HH611 pKa = 5.28ATGEE615 pKa = 4.39ITSSVGDD622 pKa = 4.01RR623 pKa = 11.84IGGIAGCNDD632 pKa = 2.93QGTFTACYY640 pKa = 8.88WEE642 pKa = 4.3NNQEE646 pKa = 3.97QGTGSGSTGGTAKK659 pKa = 10.75VEE661 pKa = 4.49GNVIWKK667 pKa = 8.03TAVDD671 pKa = 3.78AMNAALNKK679 pKa = 10.53SGWTYY684 pKa = 11.48RR685 pKa = 11.84LGGGNLPEE693 pKa = 4.67LVQNQQ698 pKa = 3.27
Molecular weight: 72.42 kDa
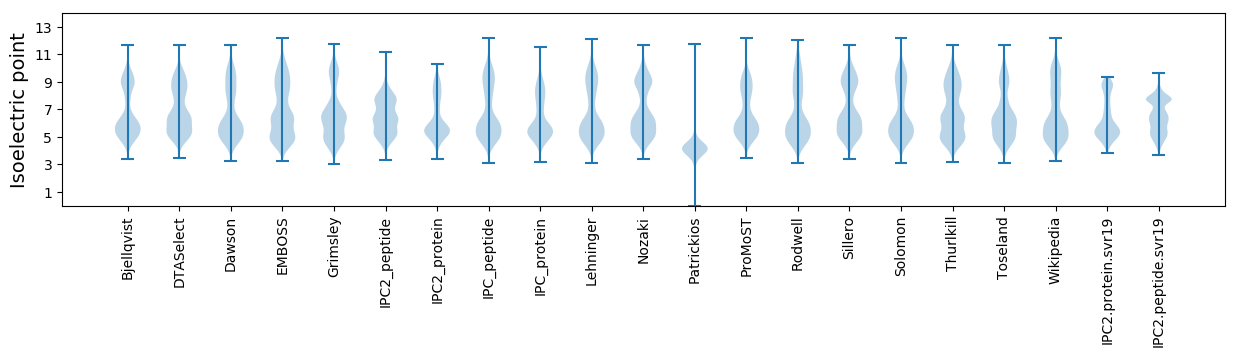
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R5C8U0|R5C8U0_9BACE Uncharacterized protein OS=Bacteroides sp. CAG:598 OX=1262743 GN=BN727_01105 PE=4 SV=1
MM1 pKa = 7.11FFARR5 pKa = 11.84FVLSLLRR12 pKa = 11.84ITLIRR17 pKa = 11.84QHH19 pKa = 6.19MNNPHH24 pKa = 6.1ILGTEE29 pKa = 4.43RR30 pKa = 11.84IGKK33 pKa = 9.35LLVQYY38 pKa = 8.77SIPAIISMTIVSLYY52 pKa = 10.82NIIDD56 pKa = 4.03SIFIGHH62 pKa = 6.59GVGPMAIAGLAITFPLMNLTAAFCNLVSAGGSTISSIRR100 pKa = 11.84LGQKK104 pKa = 10.49DD105 pKa = 3.62MEE107 pKa = 4.66GVNKK111 pKa = 10.05VLGNTLMLCLINAVVFGGLTFIFLDD136 pKa = 4.49AILRR140 pKa = 11.84FFGASGEE147 pKa = 4.3TLPYY151 pKa = 10.71ARR153 pKa = 11.84DD154 pKa = 3.62FMQVILLGTPITFTMIGLNHH174 pKa = 5.57VMRR177 pKa = 11.84ATGHH181 pKa = 6.35PKK183 pKa = 10.28KK184 pKa = 10.86AMLTSLTTVVCNIILAPIFIFQFGWGIRR212 pKa = 11.84GAALATVISQFIGMLWVLNHH232 pKa = 6.35FLQRR236 pKa = 11.84SNPIRR241 pKa = 11.84LRR243 pKa = 11.84HH244 pKa = 5.28GFQRR248 pKa = 11.84MEE250 pKa = 3.66RR251 pKa = 11.84RR252 pKa = 11.84IVGSIFSIGMSPFLMNVTACAIVIIINNSLQRR284 pKa = 11.84HH285 pKa = 5.66GGDD288 pKa = 3.4MAIGAYY294 pKa = 10.19GIINRR299 pKa = 11.84LLMLYY304 pKa = 10.47VMVVMGLTMGMQPITGYY321 pKa = 10.84NFGARR326 pKa = 11.84RR327 pKa = 11.84LDD329 pKa = 3.25RR330 pKa = 11.84VKK332 pKa = 10.47QVLRR336 pKa = 11.84LGITAGVCITSSGFLICEE354 pKa = 4.72LFPHH358 pKa = 6.83AVASVFTSDD367 pKa = 3.07EE368 pKa = 3.84ALINMAARR376 pKa = 11.84GVRR379 pKa = 11.84ICVAIFPFVGAQIVIGNFFQSIGMAKK405 pKa = 9.95ISIFMSLTRR414 pKa = 11.84QLLYY418 pKa = 10.91LLPCLLVFPQFMGLDD433 pKa = 4.67GIWTSMPVSDD443 pKa = 5.14FFAFVTAVLILLWYY457 pKa = 9.82VRR459 pKa = 11.84RR460 pKa = 11.84KK461 pKa = 10.1HH462 pKa = 6.28LFHH465 pKa = 7.01PQAIQQ470 pKa = 2.88
MM1 pKa = 7.11FFARR5 pKa = 11.84FVLSLLRR12 pKa = 11.84ITLIRR17 pKa = 11.84QHH19 pKa = 6.19MNNPHH24 pKa = 6.1ILGTEE29 pKa = 4.43RR30 pKa = 11.84IGKK33 pKa = 9.35LLVQYY38 pKa = 8.77SIPAIISMTIVSLYY52 pKa = 10.82NIIDD56 pKa = 4.03SIFIGHH62 pKa = 6.59GVGPMAIAGLAITFPLMNLTAAFCNLVSAGGSTISSIRR100 pKa = 11.84LGQKK104 pKa = 10.49DD105 pKa = 3.62MEE107 pKa = 4.66GVNKK111 pKa = 10.05VLGNTLMLCLINAVVFGGLTFIFLDD136 pKa = 4.49AILRR140 pKa = 11.84FFGASGEE147 pKa = 4.3TLPYY151 pKa = 10.71ARR153 pKa = 11.84DD154 pKa = 3.62FMQVILLGTPITFTMIGLNHH174 pKa = 5.57VMRR177 pKa = 11.84ATGHH181 pKa = 6.35PKK183 pKa = 10.28KK184 pKa = 10.86AMLTSLTTVVCNIILAPIFIFQFGWGIRR212 pKa = 11.84GAALATVISQFIGMLWVLNHH232 pKa = 6.35FLQRR236 pKa = 11.84SNPIRR241 pKa = 11.84LRR243 pKa = 11.84HH244 pKa = 5.28GFQRR248 pKa = 11.84MEE250 pKa = 3.66RR251 pKa = 11.84RR252 pKa = 11.84IVGSIFSIGMSPFLMNVTACAIVIIINNSLQRR284 pKa = 11.84HH285 pKa = 5.66GGDD288 pKa = 3.4MAIGAYY294 pKa = 10.19GIINRR299 pKa = 11.84LLMLYY304 pKa = 10.47VMVVMGLTMGMQPITGYY321 pKa = 10.84NFGARR326 pKa = 11.84RR327 pKa = 11.84LDD329 pKa = 3.25RR330 pKa = 11.84VKK332 pKa = 10.47QVLRR336 pKa = 11.84LGITAGVCITSSGFLICEE354 pKa = 4.72LFPHH358 pKa = 6.83AVASVFTSDD367 pKa = 3.07EE368 pKa = 3.84ALINMAARR376 pKa = 11.84GVRR379 pKa = 11.84ICVAIFPFVGAQIVIGNFFQSIGMAKK405 pKa = 9.95ISIFMSLTRR414 pKa = 11.84QLLYY418 pKa = 10.91LLPCLLVFPQFMGLDD433 pKa = 4.67GIWTSMPVSDD443 pKa = 5.14FFAFVTAVLILLWYY457 pKa = 9.82VRR459 pKa = 11.84RR460 pKa = 11.84KK461 pKa = 10.1HH462 pKa = 6.28LFHH465 pKa = 7.01PQAIQQ470 pKa = 2.88
Molecular weight: 51.81 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
985690 |
29 |
2799 |
355.1 |
39.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.682 ± 0.05 | 1.329 ± 0.017 |
5.656 ± 0.036 | 6.512 ± 0.042 |
4.386 ± 0.032 | 7.042 ± 0.047 |
1.994 ± 0.022 | 6.2 ± 0.045 |
5.838 ± 0.036 | 9.547 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.778 ± 0.02 | 4.582 ± 0.037 |
3.85 ± 0.027 | 3.645 ± 0.029 |
5.207 ± 0.037 | 5.701 ± 0.034 |
5.667 ± 0.036 | 6.741 ± 0.037 |
1.311 ± 0.02 | 4.325 ± 0.035 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
