
Methanophagales archaeon
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Methanomicrobia; Methanophagales; unclassified Methanophagales
Average proteome isoelectric point is 6.48
Get precalculated fractions of proteins
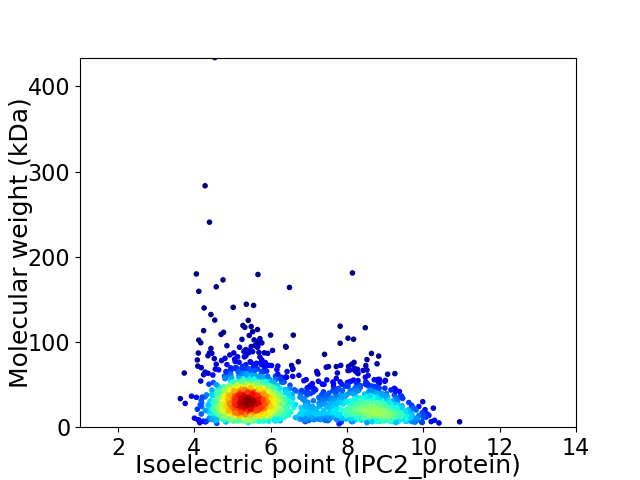
Virtual 2D-PAGE plot for 1797 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
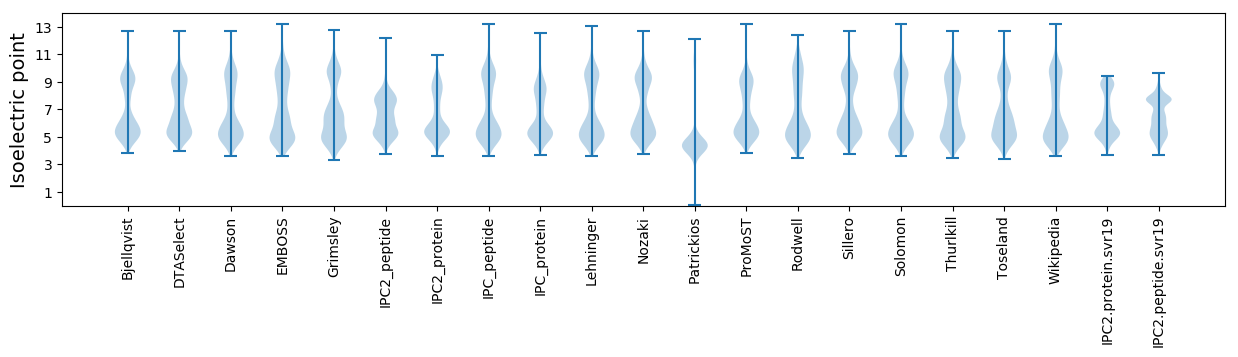
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2V3JGB0|A0A2V3JGB0_9EURY Uncharacterized protein OS=Methanophagales archaeon OX=2056316 GN=C4B56_04215 PE=4 SV=1
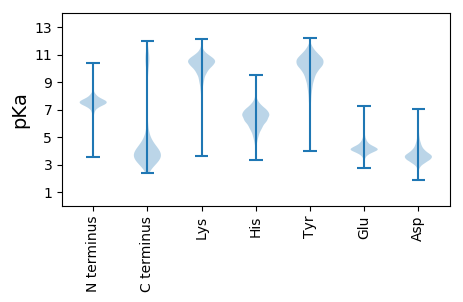
MM1 pKa = 7.25EE2 pKa = 5.67KK3 pKa = 10.82KK4 pKa = 9.31MFKK7 pKa = 10.14PNEE10 pKa = 3.72KK11 pKa = 9.54RR12 pKa = 11.84VCVLRR17 pKa = 11.84NVGIILVGVVVLSLLVCAASTATWTVSEE45 pKa = 4.15EE46 pKa = 3.99GGAGFMRR53 pKa = 11.84IEE55 pKa = 4.47DD56 pKa = 3.8AVVAASSGDD65 pKa = 3.83IIDD68 pKa = 4.83KK69 pKa = 10.95DD70 pKa = 4.3LAQTDD75 pKa = 3.62AKK77 pKa = 11.0LITTLSGEE85 pKa = 4.14AMDD88 pKa = 5.31DD89 pKa = 3.56RR90 pKa = 11.84LHH92 pKa = 5.99ITPSNVNGDD101 pKa = 4.14VNNDD105 pKa = 3.19GYY107 pKa = 11.62DD108 pKa = 3.79DD109 pKa = 4.12VVSGAWGNDD118 pKa = 2.96AGGTNAGRR126 pKa = 11.84VYY128 pKa = 10.2IYY130 pKa = 10.72LGGSTMDD137 pKa = 3.49NSSDD141 pKa = 3.71IIFTGEE147 pKa = 3.68AANDD151 pKa = 3.76EE152 pKa = 4.8LGDD155 pKa = 3.74PAAFAGDD162 pKa = 4.02VNNDD166 pKa = 3.13GYY168 pKa = 11.61DD169 pKa = 3.63DD170 pKa = 4.6VIAGAPYY177 pKa = 10.66NDD179 pKa = 4.68AGGADD184 pKa = 3.58AGRR187 pKa = 11.84AYY189 pKa = 9.85ILFGGNPMDD198 pKa = 5.48NIPDD202 pKa = 3.85VIITGQSPGDD212 pKa = 3.57NLGAWGATYY221 pKa = 10.7AGDD224 pKa = 3.75VNKK227 pKa = 10.9DD228 pKa = 3.14GFDD231 pKa = 3.71DD232 pKa = 4.79VIVSACYY239 pKa = 9.78KK240 pKa = 10.08GPGYY244 pKa = 10.92AYY246 pKa = 9.38IYY248 pKa = 10.55LGGNPMDD255 pKa = 4.02NVSDD259 pKa = 3.86LVLSGEE265 pKa = 4.34STGDD269 pKa = 3.47QFGYY273 pKa = 7.78WCAYY277 pKa = 10.15AGDD280 pKa = 4.19INKK283 pKa = 10.28DD284 pKa = 3.24GYY286 pKa = 11.49DD287 pKa = 3.73DD288 pKa = 4.78VIVGAPRR295 pKa = 11.84YY296 pKa = 10.32NGDD299 pKa = 3.05TGRR302 pKa = 11.84AYY304 pKa = 9.54IYY306 pKa = 10.65FGGKK310 pKa = 9.55PMDD313 pKa = 3.69TTPDD317 pKa = 3.51VILTGSGANHH327 pKa = 6.98KK328 pKa = 9.97FGSVGSAGDD337 pKa = 3.81VNNDD341 pKa = 3.34GYY343 pKa = 10.45PDD345 pKa = 3.86VVVGAHH351 pKa = 6.57RR352 pKa = 11.84SDD354 pKa = 2.96MAYY357 pKa = 9.67IFFGGDD363 pKa = 3.22SMDD366 pKa = 3.45NTADD370 pKa = 3.18VTMSGEE376 pKa = 4.12SSGSFFGYY384 pKa = 9.8GVSSIGDD391 pKa = 3.64VNKK394 pKa = 10.65DD395 pKa = 3.15GYY397 pKa = 11.23SDD399 pKa = 4.01VIIGAPCYY407 pKa = 10.13SDD409 pKa = 3.37SGKK412 pKa = 10.9AYY414 pKa = 9.52IYY416 pKa = 10.7FGGNPMDD423 pKa = 4.54NNPDD427 pKa = 3.0ITITGSSGDD436 pKa = 3.8GFGMTVGFAGDD447 pKa = 3.56VNGDD451 pKa = 3.6GNNDD455 pKa = 3.39FTIGAPQNNINGKK468 pKa = 9.23GRR470 pKa = 11.84TYY472 pKa = 10.37VYY474 pKa = 10.06TVISEE479 pKa = 4.42STFPVHH485 pKa = 6.72NLNTGEE491 pKa = 4.12DD492 pKa = 3.91FEE494 pKa = 5.29TIQEE498 pKa = 4.76AIDD501 pKa = 5.49DD502 pKa = 4.6PDD504 pKa = 3.74TKK506 pKa = 10.69NGHH509 pKa = 5.76TIIVDD514 pKa = 3.15AGTYY518 pKa = 9.05YY519 pKa = 11.05EE520 pKa = 3.99NVVVNKK526 pKa = 10.42SLTLKK531 pKa = 10.53GIGMPVVDD539 pKa = 4.65AGGSGSAITLSADD552 pKa = 4.08GITLEE557 pKa = 5.13GFTATNSGSSYY568 pKa = 9.96PDD570 pKa = 2.99AGIKK574 pKa = 9.99INSNNNAIMDD584 pKa = 3.67NTVCNNNYY592 pKa = 10.06GGIWLSEE599 pKa = 3.93SSNNNLQPTSGIPLL613 pKa = 3.71
MM1 pKa = 7.25EE2 pKa = 5.67KK3 pKa = 10.82KK4 pKa = 9.31MFKK7 pKa = 10.14PNEE10 pKa = 3.72KK11 pKa = 9.54RR12 pKa = 11.84VCVLRR17 pKa = 11.84NVGIILVGVVVLSLLVCAASTATWTVSEE45 pKa = 4.15EE46 pKa = 3.99GGAGFMRR53 pKa = 11.84IEE55 pKa = 4.47DD56 pKa = 3.8AVVAASSGDD65 pKa = 3.83IIDD68 pKa = 4.83KK69 pKa = 10.95DD70 pKa = 4.3LAQTDD75 pKa = 3.62AKK77 pKa = 11.0LITTLSGEE85 pKa = 4.14AMDD88 pKa = 5.31DD89 pKa = 3.56RR90 pKa = 11.84LHH92 pKa = 5.99ITPSNVNGDD101 pKa = 4.14VNNDD105 pKa = 3.19GYY107 pKa = 11.62DD108 pKa = 3.79DD109 pKa = 4.12VVSGAWGNDD118 pKa = 2.96AGGTNAGRR126 pKa = 11.84VYY128 pKa = 10.2IYY130 pKa = 10.72LGGSTMDD137 pKa = 3.49NSSDD141 pKa = 3.71IIFTGEE147 pKa = 3.68AANDD151 pKa = 3.76EE152 pKa = 4.8LGDD155 pKa = 3.74PAAFAGDD162 pKa = 4.02VNNDD166 pKa = 3.13GYY168 pKa = 11.61DD169 pKa = 3.63DD170 pKa = 4.6VIAGAPYY177 pKa = 10.66NDD179 pKa = 4.68AGGADD184 pKa = 3.58AGRR187 pKa = 11.84AYY189 pKa = 9.85ILFGGNPMDD198 pKa = 5.48NIPDD202 pKa = 3.85VIITGQSPGDD212 pKa = 3.57NLGAWGATYY221 pKa = 10.7AGDD224 pKa = 3.75VNKK227 pKa = 10.9DD228 pKa = 3.14GFDD231 pKa = 3.71DD232 pKa = 4.79VIVSACYY239 pKa = 9.78KK240 pKa = 10.08GPGYY244 pKa = 10.92AYY246 pKa = 9.38IYY248 pKa = 10.55LGGNPMDD255 pKa = 4.02NVSDD259 pKa = 3.86LVLSGEE265 pKa = 4.34STGDD269 pKa = 3.47QFGYY273 pKa = 7.78WCAYY277 pKa = 10.15AGDD280 pKa = 4.19INKK283 pKa = 10.28DD284 pKa = 3.24GYY286 pKa = 11.49DD287 pKa = 3.73DD288 pKa = 4.78VIVGAPRR295 pKa = 11.84YY296 pKa = 10.32NGDD299 pKa = 3.05TGRR302 pKa = 11.84AYY304 pKa = 9.54IYY306 pKa = 10.65FGGKK310 pKa = 9.55PMDD313 pKa = 3.69TTPDD317 pKa = 3.51VILTGSGANHH327 pKa = 6.98KK328 pKa = 9.97FGSVGSAGDD337 pKa = 3.81VNNDD341 pKa = 3.34GYY343 pKa = 10.45PDD345 pKa = 3.86VVVGAHH351 pKa = 6.57RR352 pKa = 11.84SDD354 pKa = 2.96MAYY357 pKa = 9.67IFFGGDD363 pKa = 3.22SMDD366 pKa = 3.45NTADD370 pKa = 3.18VTMSGEE376 pKa = 4.12SSGSFFGYY384 pKa = 9.8GVSSIGDD391 pKa = 3.64VNKK394 pKa = 10.65DD395 pKa = 3.15GYY397 pKa = 11.23SDD399 pKa = 4.01VIIGAPCYY407 pKa = 10.13SDD409 pKa = 3.37SGKK412 pKa = 10.9AYY414 pKa = 9.52IYY416 pKa = 10.7FGGNPMDD423 pKa = 4.54NNPDD427 pKa = 3.0ITITGSSGDD436 pKa = 3.8GFGMTVGFAGDD447 pKa = 3.56VNGDD451 pKa = 3.6GNNDD455 pKa = 3.39FTIGAPQNNINGKK468 pKa = 9.23GRR470 pKa = 11.84TYY472 pKa = 10.37VYY474 pKa = 10.06TVISEE479 pKa = 4.42STFPVHH485 pKa = 6.72NLNTGEE491 pKa = 4.12DD492 pKa = 3.91FEE494 pKa = 5.29TIQEE498 pKa = 4.76AIDD501 pKa = 5.49DD502 pKa = 4.6PDD504 pKa = 3.74TKK506 pKa = 10.69NGHH509 pKa = 5.76TIIVDD514 pKa = 3.15AGTYY518 pKa = 9.05YY519 pKa = 11.05EE520 pKa = 3.99NVVVNKK526 pKa = 10.42SLTLKK531 pKa = 10.53GIGMPVVDD539 pKa = 4.65AGGSGSAITLSADD552 pKa = 4.08GITLEE557 pKa = 5.13GFTATNSGSSYY568 pKa = 9.96PDD570 pKa = 2.99AGIKK574 pKa = 9.99INSNNNAIMDD584 pKa = 3.67NTVCNNNYY592 pKa = 10.06GGIWLSEE599 pKa = 3.93SSNNNLQPTSGIPLL613 pKa = 3.71
Molecular weight: 63.59 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2V3J810|A0A2V3J810_9EURY Radical SAM protein OS=Methanophagales archaeon OX=2056316 GN=C4B56_08085 PE=4 SV=1
MM1 pKa = 7.88AIGKK5 pKa = 6.45NTRR8 pKa = 11.84GRR10 pKa = 11.84KK11 pKa = 9.0LRR13 pKa = 11.84LCKK16 pKa = 10.37ANRR19 pKa = 11.84QNRR22 pKa = 11.84RR23 pKa = 11.84VPAWIMVKK31 pKa = 7.97TNRR34 pKa = 11.84RR35 pKa = 11.84VTTHH39 pKa = 6.29PKK41 pKa = 8.12RR42 pKa = 11.84RR43 pKa = 11.84NWRR46 pKa = 11.84RR47 pKa = 11.84TSLKK51 pKa = 10.22VV52 pKa = 3.08
MM1 pKa = 7.88AIGKK5 pKa = 6.45NTRR8 pKa = 11.84GRR10 pKa = 11.84KK11 pKa = 9.0LRR13 pKa = 11.84LCKK16 pKa = 10.37ANRR19 pKa = 11.84QNRR22 pKa = 11.84RR23 pKa = 11.84VPAWIMVKK31 pKa = 7.97TNRR34 pKa = 11.84RR35 pKa = 11.84VTTHH39 pKa = 6.29PKK41 pKa = 8.12RR42 pKa = 11.84RR43 pKa = 11.84NWRR46 pKa = 11.84RR47 pKa = 11.84TSLKK51 pKa = 10.22VV52 pKa = 3.08
Molecular weight: 6.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
535814 |
34 |
3891 |
298.2 |
33.37 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.574 ± 0.062 | 1.311 ± 0.025 |
5.281 ± 0.049 | 8.236 ± 0.075 |
3.842 ± 0.041 | 7.581 ± 0.056 |
1.846 ± 0.023 | 7.891 ± 0.047 |
6.264 ± 0.058 | 9.01 ± 0.068 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.583 ± 0.029 | 3.817 ± 0.078 |
3.94 ± 0.034 | 1.962 ± 0.028 |
6.058 ± 0.07 | 5.796 ± 0.043 |
5.02 ± 0.066 | 7.394 ± 0.046 |
1.07 ± 0.026 | 3.523 ± 0.04 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
