
Alicyclobacillus acidocaldarius subsp. acidocaldarius (strain ATCC 27009 / DSM 446 / BCRC 14685 / JCM 5260 / KCTC 1825 / NBRC 15652 / NCIMB 11725 / NRRL B-14509 / 104-IA) (Bacillus acidocaldarius)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Alicyclobacillaceae; Alicyclobacillus; Alicyclobacillus acidocaldarius; Alicyclobacillus acidocaldarius subsp. acidocaldarius
Average proteome isoelectric point is 6.57
Get precalculated fractions of proteins
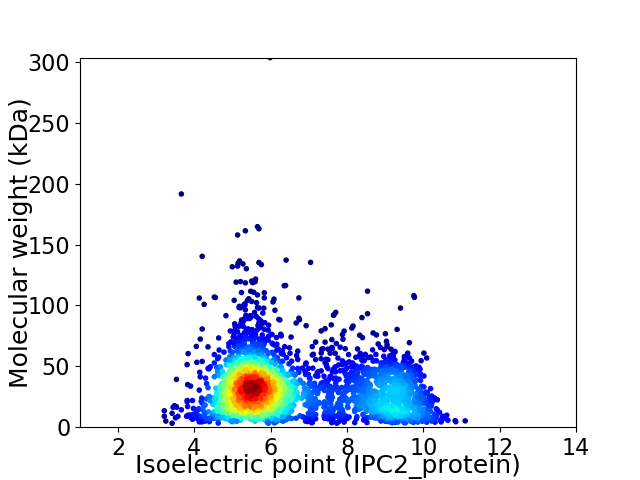
Virtual 2D-PAGE plot for 3059 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
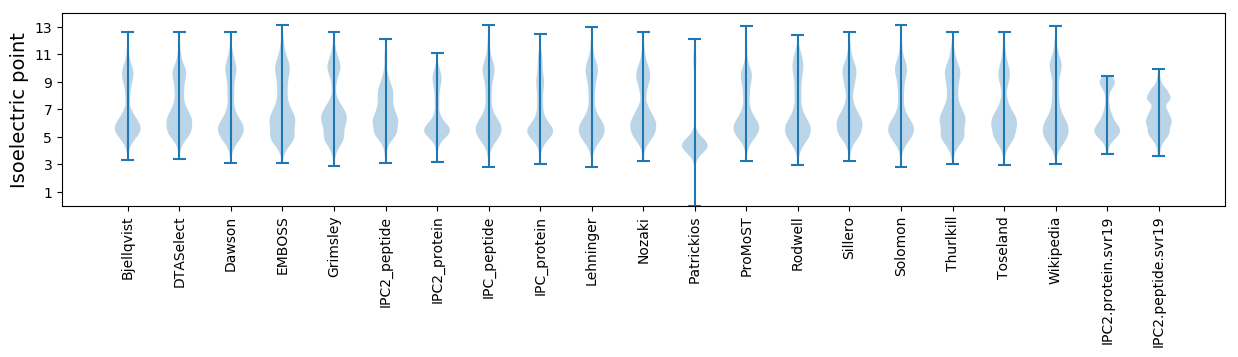
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C8WT77|C8WT77_ALIAD Uncharacterized protein OS=Alicyclobacillus acidocaldarius subsp. acidocaldarius (strain ATCC 27009 / DSM 446 / BCRC 14685 / JCM 5260 / KCTC 1825 / NBRC 15652 / NCIMB 11725 / NRRL B-14509 / 104-IA) OX=521098 GN=Aaci_2587 PE=4 SV=1
MM1 pKa = 8.12RR2 pKa = 11.84ITPWMEE8 pKa = 3.59QQQFLYY14 pKa = 11.04NMEE17 pKa = 4.96NINTAMQNVQEE28 pKa = 4.28EE29 pKa = 4.85LSTGKK34 pKa = 9.99VLNLPQDD41 pKa = 3.7DD42 pKa = 4.06PVAVSEE48 pKa = 5.35DD49 pKa = 3.42MDD51 pKa = 4.12LSAEE55 pKa = 4.17VSQVSASLSTISTGLSWMNATQTAIQGIISQLQQIQSDD93 pKa = 3.33IVAALNTPNQSQGGLAGYY111 pKa = 9.35AATVSQLIQGVYY123 pKa = 9.99QIADD127 pKa = 3.18QMVGNRR133 pKa = 11.84YY134 pKa = 9.66LFGGQADD141 pKa = 3.83NTAPTTYY148 pKa = 10.64LKK150 pKa = 9.96TSPTDD155 pKa = 3.46TPQPIPPGAGPSSGSRR171 pKa = 11.84SYY173 pKa = 11.87DD174 pKa = 3.34PFDD177 pKa = 5.41DD178 pKa = 4.4SQASPPNINYY188 pKa = 10.03QIADD192 pKa = 3.84GVSVRR197 pKa = 11.84VTVTAYY203 pKa = 10.64DD204 pKa = 3.84LFLVQPSSTAPNLQDD219 pKa = 3.43TLNNIQTDD227 pKa = 3.86IYY229 pKa = 10.67SGNSGNLKK237 pKa = 9.98QDD239 pKa = 3.57LSDD242 pKa = 3.68LQVNLNQVINLNAALGARR260 pKa = 11.84IQRR263 pKa = 11.84MTAAQNQLTQYY274 pKa = 8.03QTNLTDD280 pKa = 3.67LKK282 pKa = 11.11GGIEE286 pKa = 4.33GADD289 pKa = 3.55MARR292 pKa = 11.84VLTQFSTDD300 pKa = 3.37QVIYY304 pKa = 10.24EE305 pKa = 4.53AALQMGAQILLPSLVNYY322 pKa = 10.09LPSS325 pKa = 3.43
MM1 pKa = 8.12RR2 pKa = 11.84ITPWMEE8 pKa = 3.59QQQFLYY14 pKa = 11.04NMEE17 pKa = 4.96NINTAMQNVQEE28 pKa = 4.28EE29 pKa = 4.85LSTGKK34 pKa = 9.99VLNLPQDD41 pKa = 3.7DD42 pKa = 4.06PVAVSEE48 pKa = 5.35DD49 pKa = 3.42MDD51 pKa = 4.12LSAEE55 pKa = 4.17VSQVSASLSTISTGLSWMNATQTAIQGIISQLQQIQSDD93 pKa = 3.33IVAALNTPNQSQGGLAGYY111 pKa = 9.35AATVSQLIQGVYY123 pKa = 9.99QIADD127 pKa = 3.18QMVGNRR133 pKa = 11.84YY134 pKa = 9.66LFGGQADD141 pKa = 3.83NTAPTTYY148 pKa = 10.64LKK150 pKa = 9.96TSPTDD155 pKa = 3.46TPQPIPPGAGPSSGSRR171 pKa = 11.84SYY173 pKa = 11.87DD174 pKa = 3.34PFDD177 pKa = 5.41DD178 pKa = 4.4SQASPPNINYY188 pKa = 10.03QIADD192 pKa = 3.84GVSVRR197 pKa = 11.84VTVTAYY203 pKa = 10.64DD204 pKa = 3.84LFLVQPSSTAPNLQDD219 pKa = 3.43TLNNIQTDD227 pKa = 3.86IYY229 pKa = 10.67SGNSGNLKK237 pKa = 9.98QDD239 pKa = 3.57LSDD242 pKa = 3.68LQVNLNQVINLNAALGARR260 pKa = 11.84IQRR263 pKa = 11.84MTAAQNQLTQYY274 pKa = 8.03QTNLTDD280 pKa = 3.67LKK282 pKa = 11.11GGIEE286 pKa = 4.33GADD289 pKa = 3.55MARR292 pKa = 11.84VLTQFSTDD300 pKa = 3.37QVIYY304 pKa = 10.24EE305 pKa = 4.53AALQMGAQILLPSLVNYY322 pKa = 10.09LPSS325 pKa = 3.43
Molecular weight: 34.97 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C8WVG3|C8WVG3_ALIAD NUDIX hydrolase OS=Alicyclobacillus acidocaldarius subsp. acidocaldarius (strain ATCC 27009 / DSM 446 / BCRC 14685 / JCM 5260 / KCTC 1825 / NBRC 15652 / NCIMB 11725 / NRRL B-14509 / 104-IA) OX=521098 GN=Aaci_1047 PE=3 SV=1
MM1 pKa = 7.52KK2 pKa = 8.55PTYY5 pKa = 9.7QPNVRR10 pKa = 11.84KK11 pKa = 9.84RR12 pKa = 11.84KK13 pKa = 8.89KK14 pKa = 8.99NHH16 pKa = 5.0GFRR19 pKa = 11.84KK20 pKa = 10.05RR21 pKa = 11.84MATKK25 pKa = 9.89GGRR28 pKa = 11.84RR29 pKa = 11.84VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.94GRR39 pKa = 11.84KK40 pKa = 8.67VLSAA44 pKa = 4.05
MM1 pKa = 7.52KK2 pKa = 8.55PTYY5 pKa = 9.7QPNVRR10 pKa = 11.84KK11 pKa = 9.84RR12 pKa = 11.84KK13 pKa = 8.89KK14 pKa = 8.99NHH16 pKa = 5.0GFRR19 pKa = 11.84KK20 pKa = 10.05RR21 pKa = 11.84MATKK25 pKa = 9.89GGRR28 pKa = 11.84RR29 pKa = 11.84VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.94GRR39 pKa = 11.84KK40 pKa = 8.67VLSAA44 pKa = 4.05
Molecular weight: 5.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
925247 |
31 |
2716 |
302.5 |
33.41 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.02 ± 0.063 | 0.893 ± 0.013 |
5.162 ± 0.033 | 6.477 ± 0.051 |
3.581 ± 0.032 | 7.778 ± 0.043 |
2.395 ± 0.019 | 4.948 ± 0.035 |
3.075 ± 0.034 | 10.002 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.605 ± 0.021 | 2.481 ± 0.035 |
5.035 ± 0.033 | 3.534 ± 0.04 |
7.667 ± 0.062 | 5.294 ± 0.033 |
4.896 ± 0.035 | 8.678 ± 0.043 |
1.627 ± 0.024 | 2.852 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
