
Enterobacteria phage f1 (Bacteriophage f1)
Taxonomy: Viruses; Monodnaviria; Loebvirae; Hofneiviricota; Faserviricetes; Tubulavirales; Inoviridae; Inovirus; Escherichia virus M13
Average proteome isoelectric point is 7.16
Get precalculated fractions of proteins
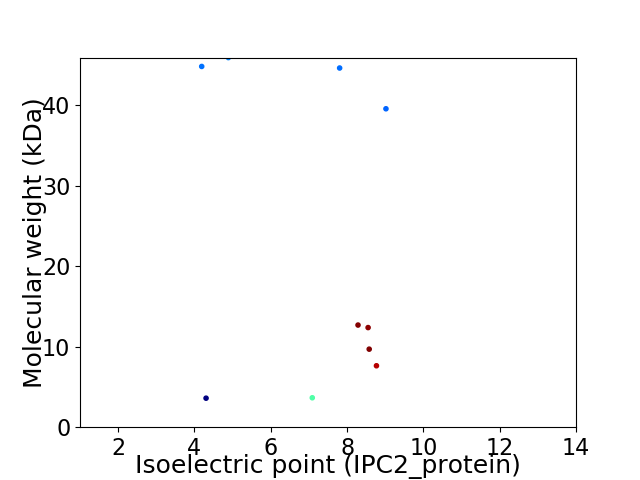
Virtual 2D-PAGE plot for 10 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
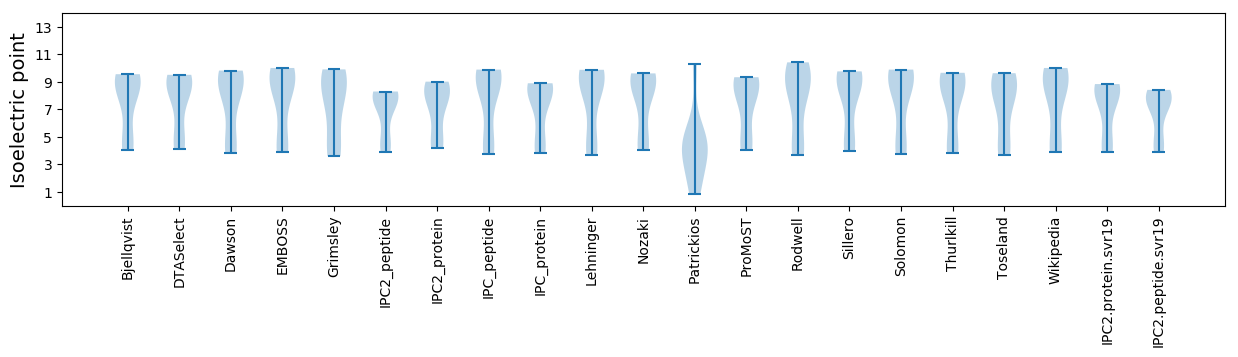
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A7BJW6|A7BJW6_BPF1 IX OS=Enterobacteria phage f1 OX=10863 GN=IX PE=4 SV=1
MM1 pKa = 7.07EE2 pKa = 4.16QVADD6 pKa = 4.42FDD8 pKa = 4.64TIYY11 pKa = 10.92QAMIQISVVLCFALGIIAGGQRR33 pKa = 3.18
MM1 pKa = 7.07EE2 pKa = 4.16QVADD6 pKa = 4.42FDD8 pKa = 4.64TIYY11 pKa = 10.92QAMIQISVVLCFALGIIAGGQRR33 pKa = 3.18
Molecular weight: 3.6 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A7BJX1|A7BJX1_BPF1 Gene 4 protein OS=Enterobacteria phage f1 OX=10863 GN=IV PE=3 SV=1
MM1 pKa = 7.44AVYY4 pKa = 9.51FVTGKK9 pKa = 10.48LGSGKK14 pKa = 8.07TLVSVGKK21 pKa = 9.86IQDD24 pKa = 4.07KK25 pKa = 10.55IVAGCKK31 pKa = 9.34IATNLDD37 pKa = 3.28LRR39 pKa = 11.84LQNLPQVGRR48 pKa = 11.84FAKK51 pKa = 9.44TPRR54 pKa = 11.84VLRR57 pKa = 11.84IPDD60 pKa = 3.64KK61 pKa = 11.0PSISDD66 pKa = 3.71LLAIGRR72 pKa = 11.84GNDD75 pKa = 3.55SYY77 pKa = 12.11DD78 pKa = 3.49EE79 pKa = 4.31NKK81 pKa = 10.66NGLLVLDD88 pKa = 4.72EE89 pKa = 4.86CGTWFNTRR97 pKa = 11.84SWNDD101 pKa = 3.27KK102 pKa = 9.17EE103 pKa = 4.09RR104 pKa = 11.84QPIIDD109 pKa = 3.83WFLHH113 pKa = 4.82ARR115 pKa = 11.84KK116 pKa = 9.63LGWDD120 pKa = 3.69IIFLVQDD127 pKa = 3.69LSIVDD132 pKa = 3.64KK133 pKa = 9.99QARR136 pKa = 11.84SALAEE141 pKa = 3.81NVVYY145 pKa = 10.39CRR147 pKa = 11.84RR148 pKa = 11.84LDD150 pKa = 4.36RR151 pKa = 11.84ITLPFVGTLYY161 pKa = 11.3SLITGSKK168 pKa = 8.19MPLPKK173 pKa = 10.44LHH175 pKa = 6.43VGVVKK180 pKa = 11.1YY181 pKa = 10.67GDD183 pKa = 3.9SQLSPTVEE191 pKa = 3.26RR192 pKa = 11.84WLYY195 pKa = 8.69TGKK198 pKa = 9.91NLYY201 pKa = 9.14NAYY204 pKa = 8.11DD205 pKa = 3.67TKK207 pKa = 11.05QAFSSNYY214 pKa = 9.25DD215 pKa = 3.42SGVYY219 pKa = 10.07SYY221 pKa = 8.2LTPYY225 pKa = 9.99LSHH228 pKa = 6.25GRR230 pKa = 11.84YY231 pKa = 8.62FKK233 pKa = 10.56PLNLGQKK240 pKa = 8.84MKK242 pKa = 9.46LTKK245 pKa = 9.88IYY247 pKa = 10.03LKK249 pKa = 10.39KK250 pKa = 10.56FSRR253 pKa = 11.84VLCLAIGFASAFTYY267 pKa = 10.44SYY269 pKa = 9.01ITQPKK274 pKa = 8.51PEE276 pKa = 4.15VKK278 pKa = 10.17KK279 pKa = 11.04VVSQTYY285 pKa = 10.61DD286 pKa = 3.05FDD288 pKa = 4.36KK289 pKa = 10.7FTIDD293 pKa = 3.03SSQRR297 pKa = 11.84LNLSYY302 pKa = 10.63RR303 pKa = 11.84YY304 pKa = 9.78VFKK307 pKa = 10.71DD308 pKa = 3.41SKK310 pKa = 11.38GKK312 pKa = 10.47LINSDD317 pKa = 4.1DD318 pKa = 4.08LQKK321 pKa = 10.71QGYY324 pKa = 8.1SLTYY328 pKa = 9.83IDD330 pKa = 5.34LCTVSIKK337 pKa = 10.46KK338 pKa = 10.48GNSNEE343 pKa = 3.96IVKK346 pKa = 10.79CNN348 pKa = 3.35
MM1 pKa = 7.44AVYY4 pKa = 9.51FVTGKK9 pKa = 10.48LGSGKK14 pKa = 8.07TLVSVGKK21 pKa = 9.86IQDD24 pKa = 4.07KK25 pKa = 10.55IVAGCKK31 pKa = 9.34IATNLDD37 pKa = 3.28LRR39 pKa = 11.84LQNLPQVGRR48 pKa = 11.84FAKK51 pKa = 9.44TPRR54 pKa = 11.84VLRR57 pKa = 11.84IPDD60 pKa = 3.64KK61 pKa = 11.0PSISDD66 pKa = 3.71LLAIGRR72 pKa = 11.84GNDD75 pKa = 3.55SYY77 pKa = 12.11DD78 pKa = 3.49EE79 pKa = 4.31NKK81 pKa = 10.66NGLLVLDD88 pKa = 4.72EE89 pKa = 4.86CGTWFNTRR97 pKa = 11.84SWNDD101 pKa = 3.27KK102 pKa = 9.17EE103 pKa = 4.09RR104 pKa = 11.84QPIIDD109 pKa = 3.83WFLHH113 pKa = 4.82ARR115 pKa = 11.84KK116 pKa = 9.63LGWDD120 pKa = 3.69IIFLVQDD127 pKa = 3.69LSIVDD132 pKa = 3.64KK133 pKa = 9.99QARR136 pKa = 11.84SALAEE141 pKa = 3.81NVVYY145 pKa = 10.39CRR147 pKa = 11.84RR148 pKa = 11.84LDD150 pKa = 4.36RR151 pKa = 11.84ITLPFVGTLYY161 pKa = 11.3SLITGSKK168 pKa = 8.19MPLPKK173 pKa = 10.44LHH175 pKa = 6.43VGVVKK180 pKa = 11.1YY181 pKa = 10.67GDD183 pKa = 3.9SQLSPTVEE191 pKa = 3.26RR192 pKa = 11.84WLYY195 pKa = 8.69TGKK198 pKa = 9.91NLYY201 pKa = 9.14NAYY204 pKa = 8.11DD205 pKa = 3.67TKK207 pKa = 11.05QAFSSNYY214 pKa = 9.25DD215 pKa = 3.42SGVYY219 pKa = 10.07SYY221 pKa = 8.2LTPYY225 pKa = 9.99LSHH228 pKa = 6.25GRR230 pKa = 11.84YY231 pKa = 8.62FKK233 pKa = 10.56PLNLGQKK240 pKa = 8.84MKK242 pKa = 9.46LTKK245 pKa = 9.88IYY247 pKa = 10.03LKK249 pKa = 10.39KK250 pKa = 10.56FSRR253 pKa = 11.84VLCLAIGFASAFTYY267 pKa = 10.44SYY269 pKa = 9.01ITQPKK274 pKa = 8.51PEE276 pKa = 4.15VKK278 pKa = 10.17KK279 pKa = 11.04VVSQTYY285 pKa = 10.61DD286 pKa = 3.05FDD288 pKa = 4.36KK289 pKa = 10.7FTIDD293 pKa = 3.03SSQRR297 pKa = 11.84LNLSYY302 pKa = 10.63RR303 pKa = 11.84YY304 pKa = 9.78VFKK307 pKa = 10.71DD308 pKa = 3.41SKK310 pKa = 11.38GKK312 pKa = 10.47LINSDD317 pKa = 4.1DD318 pKa = 4.08LQKK321 pKa = 10.71QGYY324 pKa = 8.1SLTYY328 pKa = 9.83IDD330 pKa = 5.34LCTVSIKK337 pKa = 10.46KK338 pKa = 10.48GNSNEE343 pKa = 3.96IVKK346 pKa = 10.79CNN348 pKa = 3.35
Molecular weight: 39.53 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2056 |
32 |
426 |
205.6 |
22.44 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.08 ± 0.937 | 1.216 ± 0.316 |
5.642 ± 0.398 | 3.599 ± 0.644 |
5.447 ± 0.362 | 7.879 ± 1.623 |
0.827 ± 0.229 | 5.593 ± 0.711 |
5.447 ± 0.872 | 9.776 ± 1.094 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.751 ± 0.347 | 5.642 ± 0.852 |
4.426 ± 0.463 | 3.94 ± 0.315 |
3.745 ± 0.496 | 10.117 ± 0.841 |
5.545 ± 0.322 | 7.636 ± 0.799 |
0.973 ± 0.198 | 4.037 ± 0.694 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
