
Influenza A virus (strain A/little yellow-shouldered bat/Guatemala/153/2009(H17N10))
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Insthoviricetes; Articulavirales; Orthomyxoviridae; Alphainfluenzavirus; Influenza A virus; H17N10 subtype
Average proteome isoelectric point is 6.58
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 10 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
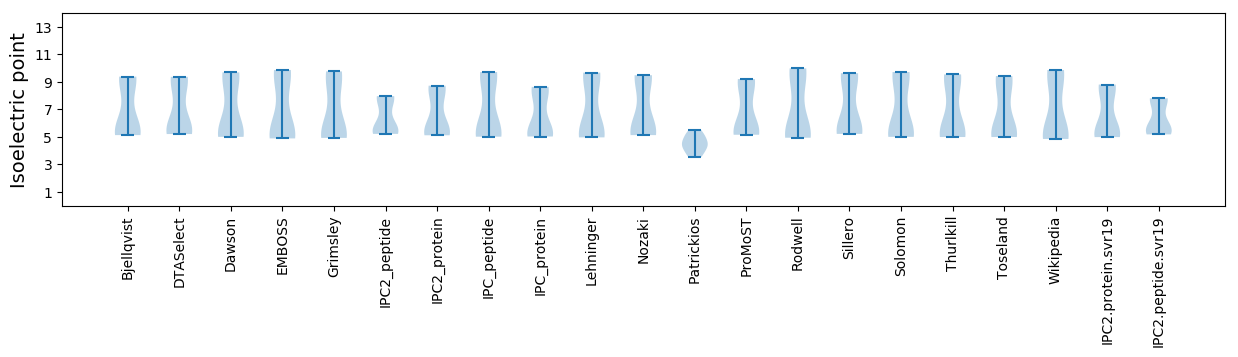
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|H6QM74|H6QM74_I09A8 Nucleoprotein OS=Influenza A virus (strain A/little yellow-shouldered bat/Guatemala/153/2009(H17N10)) OX=1129345 GN=NP PE=3 SV=1
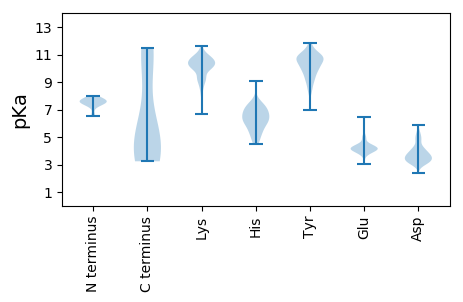
MM1 pKa = 7.41EE2 pKa = 6.43LIVLLILLNPYY13 pKa = 9.2TFVLGDD19 pKa = 4.44RR20 pKa = 11.84ICIGYY25 pKa = 8.49QANQNNQTVNTLLEE39 pKa = 4.12QNVPVTGAQEE49 pKa = 3.87ILEE52 pKa = 4.41TNHH55 pKa = 6.33NGKK58 pKa = 9.29LCSLNGVPPLDD69 pKa = 4.05LQSCTLAGWLLGNPNCDD86 pKa = 3.14SLLEE90 pKa = 4.24AEE92 pKa = 4.23EE93 pKa = 3.8WSYY96 pKa = 11.85IKK98 pKa = 10.49INEE101 pKa = 4.26SAPDD105 pKa = 3.87DD106 pKa = 3.83LCFPGNFEE114 pKa = 4.39NLQDD118 pKa = 4.78LLLEE122 pKa = 4.12MSGVQNFTKK131 pKa = 10.57VKK133 pKa = 10.12LFNPQSMTGVTTNNVDD149 pKa = 3.33QTCPFEE155 pKa = 4.68GKK157 pKa = 9.62PSFYY161 pKa = 10.79RR162 pKa = 11.84NLNWIQGNSGLPFNIEE178 pKa = 3.72IKK180 pKa = 10.87NPTSNPLLLLWGIHH194 pKa = 4.61NTKK197 pKa = 10.49DD198 pKa = 3.03AAQQRR203 pKa = 11.84NLYY206 pKa = 9.54GNDD209 pKa = 3.05YY210 pKa = 10.62SYY212 pKa = 11.22TIFNFGEE219 pKa = 4.01KK220 pKa = 9.83SEE222 pKa = 4.13EE223 pKa = 3.67FRR225 pKa = 11.84PEE227 pKa = 3.02IGQRR231 pKa = 11.84DD232 pKa = 4.06EE233 pKa = 4.83VKK235 pKa = 10.53AHH237 pKa = 6.03QDD239 pKa = 3.18RR240 pKa = 11.84IDD242 pKa = 4.02YY243 pKa = 10.38YY244 pKa = 10.08WGSLPAQSTLRR255 pKa = 11.84IEE257 pKa = 4.34STGNLIAPEE266 pKa = 3.71YY267 pKa = 9.91GFYY270 pKa = 10.8YY271 pKa = 9.98KK272 pKa = 10.4RR273 pKa = 11.84KK274 pKa = 7.93EE275 pKa = 4.23GKK277 pKa = 10.15GGLMKK282 pKa = 10.68SKK284 pKa = 10.71LPISDD289 pKa = 5.04CSTKK293 pKa = 10.63CQTPLGALNSTLPFQNVHH311 pKa = 5.06QQTIGNCPKK320 pKa = 10.16YY321 pKa = 11.03VKK323 pKa = 9.74ATSLMLATGLRR334 pKa = 11.84NNPQMEE340 pKa = 4.17GRR342 pKa = 11.84GLFGAIAGFIEE353 pKa = 5.0GGWQGMIDD361 pKa = 3.34GWYY364 pKa = 10.11GYY366 pKa = 9.32HH367 pKa = 7.04HH368 pKa = 7.2EE369 pKa = 4.18NQEE372 pKa = 4.07GSGYY376 pKa = 10.86AADD379 pKa = 5.01KK380 pKa = 10.51EE381 pKa = 4.49ATQKK385 pKa = 11.19AVDD388 pKa = 4.46AITNKK393 pKa = 9.81VNSIIDD399 pKa = 3.38KK400 pKa = 9.93MNSQFEE406 pKa = 4.63SNIKK410 pKa = 9.7EE411 pKa = 4.08FNRR414 pKa = 11.84LEE416 pKa = 4.35LRR418 pKa = 11.84IQHH421 pKa = 6.55LSDD424 pKa = 3.68RR425 pKa = 11.84VDD427 pKa = 4.29DD428 pKa = 4.57ALLDD432 pKa = 3.13IWSYY436 pKa = 8.75NTEE439 pKa = 4.04LLVLLEE445 pKa = 4.23NEE447 pKa = 4.24RR448 pKa = 11.84TLDD451 pKa = 3.57FHH453 pKa = 7.39DD454 pKa = 5.04ANVKK458 pKa = 10.21NLFEE462 pKa = 4.18KK463 pKa = 11.01VKK465 pKa = 10.96AQLKK469 pKa = 10.6DD470 pKa = 3.44NAIDD474 pKa = 3.66EE475 pKa = 4.81GNGCFLLLHH484 pKa = 6.57KK485 pKa = 10.67CNNSCMDD492 pKa = 4.78DD493 pKa = 3.59IKK495 pKa = 11.39NGTYY499 pKa = 10.34KK500 pKa = 11.01YY501 pKa = 9.21MDD503 pKa = 3.72YY504 pKa = 10.81RR505 pKa = 11.84EE506 pKa = 4.05EE507 pKa = 4.01SHH509 pKa = 7.54IEE511 pKa = 3.71KK512 pKa = 10.59QKK514 pKa = 10.49IDD516 pKa = 3.71GVKK519 pKa = 8.73LTDD522 pKa = 3.24YY523 pKa = 10.68SRR525 pKa = 11.84YY526 pKa = 10.63YY527 pKa = 10.58IMTLYY532 pKa = 9.59STIASSVVLGSLIIAAFLWGCQKK555 pKa = 11.09GSIQCKK561 pKa = 9.13ICII564 pKa = 4.08
MM1 pKa = 7.41EE2 pKa = 6.43LIVLLILLNPYY13 pKa = 9.2TFVLGDD19 pKa = 4.44RR20 pKa = 11.84ICIGYY25 pKa = 8.49QANQNNQTVNTLLEE39 pKa = 4.12QNVPVTGAQEE49 pKa = 3.87ILEE52 pKa = 4.41TNHH55 pKa = 6.33NGKK58 pKa = 9.29LCSLNGVPPLDD69 pKa = 4.05LQSCTLAGWLLGNPNCDD86 pKa = 3.14SLLEE90 pKa = 4.24AEE92 pKa = 4.23EE93 pKa = 3.8WSYY96 pKa = 11.85IKK98 pKa = 10.49INEE101 pKa = 4.26SAPDD105 pKa = 3.87DD106 pKa = 3.83LCFPGNFEE114 pKa = 4.39NLQDD118 pKa = 4.78LLLEE122 pKa = 4.12MSGVQNFTKK131 pKa = 10.57VKK133 pKa = 10.12LFNPQSMTGVTTNNVDD149 pKa = 3.33QTCPFEE155 pKa = 4.68GKK157 pKa = 9.62PSFYY161 pKa = 10.79RR162 pKa = 11.84NLNWIQGNSGLPFNIEE178 pKa = 3.72IKK180 pKa = 10.87NPTSNPLLLLWGIHH194 pKa = 4.61NTKK197 pKa = 10.49DD198 pKa = 3.03AAQQRR203 pKa = 11.84NLYY206 pKa = 9.54GNDD209 pKa = 3.05YY210 pKa = 10.62SYY212 pKa = 11.22TIFNFGEE219 pKa = 4.01KK220 pKa = 9.83SEE222 pKa = 4.13EE223 pKa = 3.67FRR225 pKa = 11.84PEE227 pKa = 3.02IGQRR231 pKa = 11.84DD232 pKa = 4.06EE233 pKa = 4.83VKK235 pKa = 10.53AHH237 pKa = 6.03QDD239 pKa = 3.18RR240 pKa = 11.84IDD242 pKa = 4.02YY243 pKa = 10.38YY244 pKa = 10.08WGSLPAQSTLRR255 pKa = 11.84IEE257 pKa = 4.34STGNLIAPEE266 pKa = 3.71YY267 pKa = 9.91GFYY270 pKa = 10.8YY271 pKa = 9.98KK272 pKa = 10.4RR273 pKa = 11.84KK274 pKa = 7.93EE275 pKa = 4.23GKK277 pKa = 10.15GGLMKK282 pKa = 10.68SKK284 pKa = 10.71LPISDD289 pKa = 5.04CSTKK293 pKa = 10.63CQTPLGALNSTLPFQNVHH311 pKa = 5.06QQTIGNCPKK320 pKa = 10.16YY321 pKa = 11.03VKK323 pKa = 9.74ATSLMLATGLRR334 pKa = 11.84NNPQMEE340 pKa = 4.17GRR342 pKa = 11.84GLFGAIAGFIEE353 pKa = 5.0GGWQGMIDD361 pKa = 3.34GWYY364 pKa = 10.11GYY366 pKa = 9.32HH367 pKa = 7.04HH368 pKa = 7.2EE369 pKa = 4.18NQEE372 pKa = 4.07GSGYY376 pKa = 10.86AADD379 pKa = 5.01KK380 pKa = 10.51EE381 pKa = 4.49ATQKK385 pKa = 11.19AVDD388 pKa = 4.46AITNKK393 pKa = 9.81VNSIIDD399 pKa = 3.38KK400 pKa = 9.93MNSQFEE406 pKa = 4.63SNIKK410 pKa = 9.7EE411 pKa = 4.08FNRR414 pKa = 11.84LEE416 pKa = 4.35LRR418 pKa = 11.84IQHH421 pKa = 6.55LSDD424 pKa = 3.68RR425 pKa = 11.84VDD427 pKa = 4.29DD428 pKa = 4.57ALLDD432 pKa = 3.13IWSYY436 pKa = 8.75NTEE439 pKa = 4.04LLVLLEE445 pKa = 4.23NEE447 pKa = 4.24RR448 pKa = 11.84TLDD451 pKa = 3.57FHH453 pKa = 7.39DD454 pKa = 5.04ANVKK458 pKa = 10.21NLFEE462 pKa = 4.18KK463 pKa = 11.01VKK465 pKa = 10.96AQLKK469 pKa = 10.6DD470 pKa = 3.44NAIDD474 pKa = 3.66EE475 pKa = 4.81GNGCFLLLHH484 pKa = 6.57KK485 pKa = 10.67CNNSCMDD492 pKa = 4.78DD493 pKa = 3.59IKK495 pKa = 11.39NGTYY499 pKa = 10.34KK500 pKa = 11.01YY501 pKa = 9.21MDD503 pKa = 3.72YY504 pKa = 10.81RR505 pKa = 11.84EE506 pKa = 4.05EE507 pKa = 4.01SHH509 pKa = 7.54IEE511 pKa = 3.71KK512 pKa = 10.59QKK514 pKa = 10.49IDD516 pKa = 3.71GVKK519 pKa = 8.73LTDD522 pKa = 3.24YY523 pKa = 10.68SRR525 pKa = 11.84YY526 pKa = 10.63YY527 pKa = 10.58IMTLYY532 pKa = 9.59STIASSVVLGSLIIAAFLWGCQKK555 pKa = 11.09GSIQCKK561 pKa = 9.13ICII564 pKa = 4.08
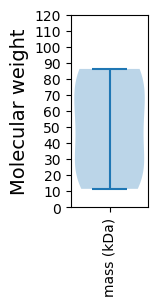
Molecular weight: 63.53 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|H6QM71|H6QM71_I09A8 RNA-directed RNA polymerase catalytic subunit OS=Influenza A virus (strain A/little yellow-shouldered bat/Guatemala/153/2009(H17N10)) OX=1129345 GN=PB1 PE=4 SV=1
MM1 pKa = 7.33EE2 pKa = 6.17RR3 pKa = 11.84IKK5 pKa = 10.93EE6 pKa = 3.92LLEE9 pKa = 3.81MVKK12 pKa = 10.28NSRR15 pKa = 11.84MRR17 pKa = 11.84EE18 pKa = 3.86ILTTTSVDD26 pKa = 2.85HH27 pKa = 5.96MAVIKK32 pKa = 10.36KK33 pKa = 7.36YY34 pKa = 9.12TSGRR38 pKa = 11.84QEE40 pKa = 3.99KK41 pKa = 10.64NPALRR46 pKa = 11.84MKK48 pKa = 10.19WMMAMKK54 pKa = 10.18YY55 pKa = 9.37PISASSRR62 pKa = 11.84IRR64 pKa = 11.84EE65 pKa = 4.21MIPEE69 pKa = 4.19KK70 pKa = 11.09DD71 pKa = 3.27EE72 pKa = 5.33DD73 pKa = 5.52GNTLWTNTKK82 pKa = 10.09DD83 pKa = 3.22AGSNRR88 pKa = 11.84VLVSPNAVTWWNRR101 pKa = 11.84AGPVSDD107 pKa = 3.54VVHH110 pKa = 6.0YY111 pKa = 9.76PRR113 pKa = 11.84VYY115 pKa = 11.01KK116 pKa = 10.19MYY118 pKa = 10.25FDD120 pKa = 3.62RR121 pKa = 11.84LEE123 pKa = 4.26RR124 pKa = 11.84LTHH127 pKa = 5.26GTFGPVKK134 pKa = 10.1FYY136 pKa = 11.42NQVKK140 pKa = 8.71VRR142 pKa = 11.84KK143 pKa = 9.41RR144 pKa = 11.84VDD146 pKa = 2.96INPGHH151 pKa = 7.14KK152 pKa = 10.4DD153 pKa = 3.12LTSRR157 pKa = 11.84EE158 pKa = 4.1AQEE161 pKa = 4.57VIMEE165 pKa = 4.2VVFPNEE171 pKa = 3.32VGARR175 pKa = 11.84TLSSDD180 pKa = 3.38AQLTITKK187 pKa = 9.41EE188 pKa = 4.04KK189 pKa = 10.8KK190 pKa = 10.23EE191 pKa = 3.87EE192 pKa = 3.95LKK194 pKa = 10.74NCKK197 pKa = 9.41ISPIMVAYY205 pKa = 8.87MLEE208 pKa = 4.23RR209 pKa = 11.84EE210 pKa = 4.37LVRR213 pKa = 11.84RR214 pKa = 11.84TRR216 pKa = 11.84FLPIAGATSSTYY228 pKa = 10.78VEE230 pKa = 4.48VLHH233 pKa = 6.22LTQGTCWEE241 pKa = 4.07QQYY244 pKa = 8.58TPGGEE249 pKa = 4.19AEE251 pKa = 5.25NDD253 pKa = 3.85DD254 pKa = 4.81LDD256 pKa = 3.62QTLIIASRR264 pKa = 11.84NIVRR268 pKa = 11.84RR269 pKa = 11.84SIVAIDD275 pKa = 3.83PLASLLSMCHH285 pKa = 4.51TTSISSEE292 pKa = 3.89PLVEE296 pKa = 4.06ILRR299 pKa = 11.84SNPTDD304 pKa = 3.37EE305 pKa = 4.35QAVNICKK312 pKa = 10.0AALGIRR318 pKa = 11.84INNSFSFGGYY328 pKa = 7.5NFKK331 pKa = 10.39RR332 pKa = 11.84VKK334 pKa = 10.43GSSQRR339 pKa = 11.84TEE341 pKa = 3.87KK342 pKa = 10.91AVLTGNLQTLTMTIFEE358 pKa = 4.88GYY360 pKa = 10.56EE361 pKa = 3.77EE362 pKa = 4.93FNVSGKK368 pKa = 9.99RR369 pKa = 11.84ASAVLKK375 pKa = 10.67KK376 pKa = 9.67GTQRR380 pKa = 11.84LIQAIIGGRR389 pKa = 11.84TLEE392 pKa = 5.21DD393 pKa = 3.46ILNLMITLMVFSQEE407 pKa = 3.89EE408 pKa = 4.23KK409 pKa = 9.24MLKK412 pKa = 9.98AVRR415 pKa = 11.84GDD417 pKa = 3.59LNFVNRR423 pKa = 11.84ANQRR427 pKa = 11.84LNPMYY432 pKa = 10.24QLLRR436 pKa = 11.84HH437 pKa = 5.39FQKK440 pKa = 10.91DD441 pKa = 3.04SSTLLRR447 pKa = 11.84NWGTEE452 pKa = 3.95EE453 pKa = 4.2IDD455 pKa = 4.31PIMGIAGIMPDD466 pKa = 3.17GTINKK471 pKa = 8.25NQTLIGVRR479 pKa = 11.84LSQGGVDD486 pKa = 3.01EE487 pKa = 4.71YY488 pKa = 11.65SFNEE492 pKa = 4.33RR493 pKa = 11.84IRR495 pKa = 11.84VNIDD499 pKa = 2.37KK500 pKa = 10.45YY501 pKa = 11.09LRR503 pKa = 11.84VRR505 pKa = 11.84NEE507 pKa = 3.58KK508 pKa = 10.96GEE510 pKa = 4.15LLISPEE516 pKa = 4.37EE517 pKa = 3.87VSEE520 pKa = 4.16AQGQEE525 pKa = 4.05KK526 pKa = 10.84LPINYY531 pKa = 9.2NSSLMWEE538 pKa = 4.36VNGPEE543 pKa = 4.26SILTNTYY550 pKa = 9.85HH551 pKa = 7.33WIIKK555 pKa = 9.32NWEE558 pKa = 4.05LLKK561 pKa = 9.1TQWMTDD567 pKa = 3.0PTVLYY572 pKa = 10.96NRR574 pKa = 11.84MEE576 pKa = 4.14FEE578 pKa = 4.45PFQTLIPKK586 pKa = 10.04GNRR589 pKa = 11.84ATYY592 pKa = 10.54SGFTRR597 pKa = 11.84TLFQQMRR604 pKa = 11.84DD605 pKa = 3.39VEE607 pKa = 4.57GTFDD611 pKa = 4.22SIQVIKK617 pKa = 10.63LLPFSAHH624 pKa = 6.46PPSLGRR630 pKa = 11.84TQFSSFTLNIRR641 pKa = 11.84GAPLRR646 pKa = 11.84LLIRR650 pKa = 11.84GNSQVFNYY658 pKa = 10.42NQMEE662 pKa = 4.12NVIIVLGKK670 pKa = 10.2SVGSPEE676 pKa = 4.2RR677 pKa = 11.84SILTEE682 pKa = 3.77SSSIEE687 pKa = 4.15SAVLRR692 pKa = 11.84GFLILGKK699 pKa = 10.26ANSKK703 pKa = 9.91YY704 pKa = 10.93GPVLTIGEE712 pKa = 4.38LDD714 pKa = 3.14KK715 pKa = 11.41LGRR718 pKa = 11.84GEE720 pKa = 4.21KK721 pKa = 10.72ANVLIGQGDD730 pKa = 3.89TVLVMKK736 pKa = 10.33RR737 pKa = 11.84KK738 pKa = 9.31RR739 pKa = 11.84DD740 pKa = 3.62SSILTDD746 pKa = 3.22SQTALKK752 pKa = 10.24RR753 pKa = 11.84IRR755 pKa = 11.84LEE757 pKa = 3.74EE758 pKa = 4.22SKK760 pKa = 11.23
MM1 pKa = 7.33EE2 pKa = 6.17RR3 pKa = 11.84IKK5 pKa = 10.93EE6 pKa = 3.92LLEE9 pKa = 3.81MVKK12 pKa = 10.28NSRR15 pKa = 11.84MRR17 pKa = 11.84EE18 pKa = 3.86ILTTTSVDD26 pKa = 2.85HH27 pKa = 5.96MAVIKK32 pKa = 10.36KK33 pKa = 7.36YY34 pKa = 9.12TSGRR38 pKa = 11.84QEE40 pKa = 3.99KK41 pKa = 10.64NPALRR46 pKa = 11.84MKK48 pKa = 10.19WMMAMKK54 pKa = 10.18YY55 pKa = 9.37PISASSRR62 pKa = 11.84IRR64 pKa = 11.84EE65 pKa = 4.21MIPEE69 pKa = 4.19KK70 pKa = 11.09DD71 pKa = 3.27EE72 pKa = 5.33DD73 pKa = 5.52GNTLWTNTKK82 pKa = 10.09DD83 pKa = 3.22AGSNRR88 pKa = 11.84VLVSPNAVTWWNRR101 pKa = 11.84AGPVSDD107 pKa = 3.54VVHH110 pKa = 6.0YY111 pKa = 9.76PRR113 pKa = 11.84VYY115 pKa = 11.01KK116 pKa = 10.19MYY118 pKa = 10.25FDD120 pKa = 3.62RR121 pKa = 11.84LEE123 pKa = 4.26RR124 pKa = 11.84LTHH127 pKa = 5.26GTFGPVKK134 pKa = 10.1FYY136 pKa = 11.42NQVKK140 pKa = 8.71VRR142 pKa = 11.84KK143 pKa = 9.41RR144 pKa = 11.84VDD146 pKa = 2.96INPGHH151 pKa = 7.14KK152 pKa = 10.4DD153 pKa = 3.12LTSRR157 pKa = 11.84EE158 pKa = 4.1AQEE161 pKa = 4.57VIMEE165 pKa = 4.2VVFPNEE171 pKa = 3.32VGARR175 pKa = 11.84TLSSDD180 pKa = 3.38AQLTITKK187 pKa = 9.41EE188 pKa = 4.04KK189 pKa = 10.8KK190 pKa = 10.23EE191 pKa = 3.87EE192 pKa = 3.95LKK194 pKa = 10.74NCKK197 pKa = 9.41ISPIMVAYY205 pKa = 8.87MLEE208 pKa = 4.23RR209 pKa = 11.84EE210 pKa = 4.37LVRR213 pKa = 11.84RR214 pKa = 11.84TRR216 pKa = 11.84FLPIAGATSSTYY228 pKa = 10.78VEE230 pKa = 4.48VLHH233 pKa = 6.22LTQGTCWEE241 pKa = 4.07QQYY244 pKa = 8.58TPGGEE249 pKa = 4.19AEE251 pKa = 5.25NDD253 pKa = 3.85DD254 pKa = 4.81LDD256 pKa = 3.62QTLIIASRR264 pKa = 11.84NIVRR268 pKa = 11.84RR269 pKa = 11.84SIVAIDD275 pKa = 3.83PLASLLSMCHH285 pKa = 4.51TTSISSEE292 pKa = 3.89PLVEE296 pKa = 4.06ILRR299 pKa = 11.84SNPTDD304 pKa = 3.37EE305 pKa = 4.35QAVNICKK312 pKa = 10.0AALGIRR318 pKa = 11.84INNSFSFGGYY328 pKa = 7.5NFKK331 pKa = 10.39RR332 pKa = 11.84VKK334 pKa = 10.43GSSQRR339 pKa = 11.84TEE341 pKa = 3.87KK342 pKa = 10.91AVLTGNLQTLTMTIFEE358 pKa = 4.88GYY360 pKa = 10.56EE361 pKa = 3.77EE362 pKa = 4.93FNVSGKK368 pKa = 9.99RR369 pKa = 11.84ASAVLKK375 pKa = 10.67KK376 pKa = 9.67GTQRR380 pKa = 11.84LIQAIIGGRR389 pKa = 11.84TLEE392 pKa = 5.21DD393 pKa = 3.46ILNLMITLMVFSQEE407 pKa = 3.89EE408 pKa = 4.23KK409 pKa = 9.24MLKK412 pKa = 9.98AVRR415 pKa = 11.84GDD417 pKa = 3.59LNFVNRR423 pKa = 11.84ANQRR427 pKa = 11.84LNPMYY432 pKa = 10.24QLLRR436 pKa = 11.84HH437 pKa = 5.39FQKK440 pKa = 10.91DD441 pKa = 3.04SSTLLRR447 pKa = 11.84NWGTEE452 pKa = 3.95EE453 pKa = 4.2IDD455 pKa = 4.31PIMGIAGIMPDD466 pKa = 3.17GTINKK471 pKa = 8.25NQTLIGVRR479 pKa = 11.84LSQGGVDD486 pKa = 3.01EE487 pKa = 4.71YY488 pKa = 11.65SFNEE492 pKa = 4.33RR493 pKa = 11.84IRR495 pKa = 11.84VNIDD499 pKa = 2.37KK500 pKa = 10.45YY501 pKa = 11.09LRR503 pKa = 11.84VRR505 pKa = 11.84NEE507 pKa = 3.58KK508 pKa = 10.96GEE510 pKa = 4.15LLISPEE516 pKa = 4.37EE517 pKa = 3.87VSEE520 pKa = 4.16AQGQEE525 pKa = 4.05KK526 pKa = 10.84LPINYY531 pKa = 9.2NSSLMWEE538 pKa = 4.36VNGPEE543 pKa = 4.26SILTNTYY550 pKa = 9.85HH551 pKa = 7.33WIIKK555 pKa = 9.32NWEE558 pKa = 4.05LLKK561 pKa = 9.1TQWMTDD567 pKa = 3.0PTVLYY572 pKa = 10.96NRR574 pKa = 11.84MEE576 pKa = 4.14FEE578 pKa = 4.45PFQTLIPKK586 pKa = 10.04GNRR589 pKa = 11.84ATYY592 pKa = 10.54SGFTRR597 pKa = 11.84TLFQQMRR604 pKa = 11.84DD605 pKa = 3.39VEE607 pKa = 4.57GTFDD611 pKa = 4.22SIQVIKK617 pKa = 10.63LLPFSAHH624 pKa = 6.46PPSLGRR630 pKa = 11.84TQFSSFTLNIRR641 pKa = 11.84GAPLRR646 pKa = 11.84LLIRR650 pKa = 11.84GNSQVFNYY658 pKa = 10.42NQMEE662 pKa = 4.12NVIIVLGKK670 pKa = 10.2SVGSPEE676 pKa = 4.2RR677 pKa = 11.84SILTEE682 pKa = 3.77SSSIEE687 pKa = 4.15SAVLRR692 pKa = 11.84GFLILGKK699 pKa = 10.26ANSKK703 pKa = 9.91YY704 pKa = 10.93GPVLTIGEE712 pKa = 4.38LDD714 pKa = 3.14KK715 pKa = 11.41LGRR718 pKa = 11.84GEE720 pKa = 4.21KK721 pKa = 10.72ANVLIGQGDD730 pKa = 3.89TVLVMKK736 pKa = 10.33RR737 pKa = 11.84KK738 pKa = 9.31RR739 pKa = 11.84DD740 pKa = 3.62SSILTDD746 pKa = 3.22SQTALKK752 pKa = 10.24RR753 pKa = 11.84IRR755 pKa = 11.84LEE757 pKa = 3.74EE758 pKa = 4.22SKK760 pKa = 11.23
Molecular weight: 86.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4423 |
96 |
760 |
442.3 |
50.19 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.63 ± 0.562 | 1.628 ± 0.315 |
4.364 ± 0.275 | 7.619 ± 0.619 |
4.002 ± 0.292 | 6.579 ± 0.587 |
1.515 ± 0.162 | 7.19 ± 0.197 |
6.308 ± 0.385 | 9.202 ± 0.477 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.482 ± 0.324 | 6.127 ± 0.509 |
3.798 ± 0.163 | 4.183 ± 0.282 |
5.811 ± 0.556 | 6.851 ± 0.34 |
6.692 ± 0.422 | 4.612 ± 0.37 |
1.515 ± 0.167 | 2.894 ± 0.251 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
