
Brachybacterium faecium (strain ATCC 43885 / DSM 4810 / JCM 11609 / LMG 19847 / NBRC 14762 / NCIMB 9860 / 6-10)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Dermabacteraceae; Brachybacterium; Brachybacterium faecium
Average proteome isoelectric point is 5.97
Get precalculated fractions of proteins
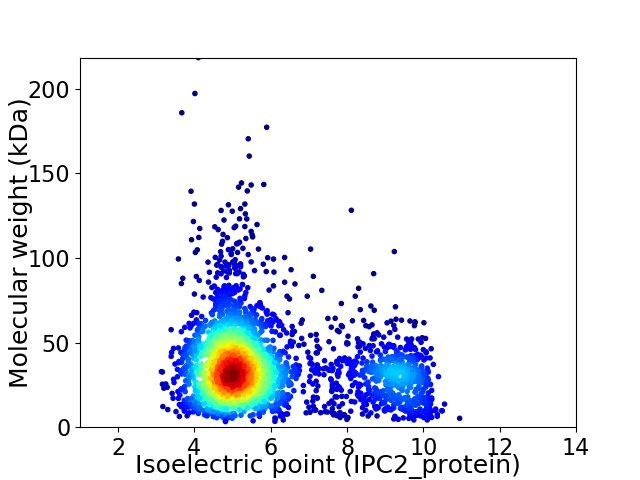
Virtual 2D-PAGE plot for 3062 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C7MHZ1|C7MHZ1_BRAFD Phosphoglycerate dehydrogenase-like oxidoreductase OS=Brachybacterium faecium (strain ATCC 43885 / DSM 4810 / JCM 11609 / LMG 19847 / NBRC 14762 / NCIMB 9860 / 6-10) OX=446465 GN=Bfae_28970 PE=3 SV=1
MM1 pKa = 7.53SSSGWGGPGNEE12 pKa = 4.15PQGGPGQQGSGQPGGPYY29 pKa = 8.24GTPSPGWQSPPPRR42 pKa = 11.84RR43 pKa = 11.84GRR45 pKa = 11.84SWALLAVAFSCVLALLLVVGGGITYY70 pKa = 10.36LALRR74 pKa = 11.84QNADD78 pKa = 3.3EE79 pKa = 4.68SSVATGSPSTSTTEE93 pKa = 4.04DD94 pKa = 3.27VSPSDD99 pKa = 3.79DD100 pKa = 3.48ASPDD104 pKa = 3.43EE105 pKa = 4.76EE106 pKa = 4.74VPPTPEE112 pKa = 3.67KK113 pKa = 11.09SEE115 pKa = 3.98TSSFEE120 pKa = 3.88VVVPYY125 pKa = 10.3DD126 pKa = 3.93PPTGTVDD133 pKa = 3.73EE134 pKa = 5.15LWDD137 pKa = 3.72VMADD141 pKa = 3.5NPLSEE146 pKa = 4.62GSLPALSTCEE156 pKa = 4.41LPATPTEE163 pKa = 4.26PDD165 pKa = 3.56DD166 pKa = 5.85AEE168 pKa = 4.23LQAVLDD174 pKa = 4.21AASTCLNQVWATASSDD190 pKa = 3.69RR191 pKa = 11.84GLPWTSPEE199 pKa = 3.47IVVYY203 pKa = 8.87HH204 pKa = 6.56HH205 pKa = 7.49PDD207 pKa = 3.13VPKK210 pKa = 10.53EE211 pKa = 4.12ATCSDD216 pKa = 3.43GFAADD221 pKa = 4.78FPRR224 pKa = 11.84VCNLDD229 pKa = 3.46STIYY233 pKa = 10.09WPDD236 pKa = 3.22GYY238 pKa = 10.64GTGRR242 pKa = 11.84DD243 pKa = 3.83LEE245 pKa = 4.59DD246 pKa = 3.89AADD249 pKa = 3.83VPAAYY254 pKa = 9.91LWDD257 pKa = 3.97LSYY260 pKa = 11.41LYY262 pKa = 10.41MNTVTWNSSLAVYY275 pKa = 9.61YY276 pKa = 9.5GTMSTQLEE284 pKa = 4.28EE285 pKa = 4.35TDD287 pKa = 5.04DD288 pKa = 3.95EE289 pKa = 4.68RR290 pKa = 11.84HH291 pKa = 5.84DD292 pKa = 4.14EE293 pKa = 3.57AWRR296 pKa = 11.84RR297 pKa = 11.84YY298 pKa = 8.77NLQNQCLASAASMQVPSAAEE318 pKa = 3.64PSKK321 pKa = 10.24QLRR324 pKa = 11.84DD325 pKa = 3.33ILTSPDD331 pKa = 2.78TWEE334 pKa = 4.5AGQPPKK340 pKa = 10.59TITPEE345 pKa = 3.83NRR347 pKa = 11.84ALWIQRR353 pKa = 11.84GFEE356 pKa = 4.03ADD358 pKa = 3.86GDD360 pKa = 4.09LSACNTWTADD370 pKa = 3.64LEE372 pKa = 4.56QVTT375 pKa = 4.3
MM1 pKa = 7.53SSSGWGGPGNEE12 pKa = 4.15PQGGPGQQGSGQPGGPYY29 pKa = 8.24GTPSPGWQSPPPRR42 pKa = 11.84RR43 pKa = 11.84GRR45 pKa = 11.84SWALLAVAFSCVLALLLVVGGGITYY70 pKa = 10.36LALRR74 pKa = 11.84QNADD78 pKa = 3.3EE79 pKa = 4.68SSVATGSPSTSTTEE93 pKa = 4.04DD94 pKa = 3.27VSPSDD99 pKa = 3.79DD100 pKa = 3.48ASPDD104 pKa = 3.43EE105 pKa = 4.76EE106 pKa = 4.74VPPTPEE112 pKa = 3.67KK113 pKa = 11.09SEE115 pKa = 3.98TSSFEE120 pKa = 3.88VVVPYY125 pKa = 10.3DD126 pKa = 3.93PPTGTVDD133 pKa = 3.73EE134 pKa = 5.15LWDD137 pKa = 3.72VMADD141 pKa = 3.5NPLSEE146 pKa = 4.62GSLPALSTCEE156 pKa = 4.41LPATPTEE163 pKa = 4.26PDD165 pKa = 3.56DD166 pKa = 5.85AEE168 pKa = 4.23LQAVLDD174 pKa = 4.21AASTCLNQVWATASSDD190 pKa = 3.69RR191 pKa = 11.84GLPWTSPEE199 pKa = 3.47IVVYY203 pKa = 8.87HH204 pKa = 6.56HH205 pKa = 7.49PDD207 pKa = 3.13VPKK210 pKa = 10.53EE211 pKa = 4.12ATCSDD216 pKa = 3.43GFAADD221 pKa = 4.78FPRR224 pKa = 11.84VCNLDD229 pKa = 3.46STIYY233 pKa = 10.09WPDD236 pKa = 3.22GYY238 pKa = 10.64GTGRR242 pKa = 11.84DD243 pKa = 3.83LEE245 pKa = 4.59DD246 pKa = 3.89AADD249 pKa = 3.83VPAAYY254 pKa = 9.91LWDD257 pKa = 3.97LSYY260 pKa = 11.41LYY262 pKa = 10.41MNTVTWNSSLAVYY275 pKa = 9.61YY276 pKa = 9.5GTMSTQLEE284 pKa = 4.28EE285 pKa = 4.35TDD287 pKa = 5.04DD288 pKa = 3.95EE289 pKa = 4.68RR290 pKa = 11.84HH291 pKa = 5.84DD292 pKa = 4.14EE293 pKa = 3.57AWRR296 pKa = 11.84RR297 pKa = 11.84YY298 pKa = 8.77NLQNQCLASAASMQVPSAAEE318 pKa = 3.64PSKK321 pKa = 10.24QLRR324 pKa = 11.84DD325 pKa = 3.33ILTSPDD331 pKa = 2.78TWEE334 pKa = 4.5AGQPPKK340 pKa = 10.59TITPEE345 pKa = 3.83NRR347 pKa = 11.84ALWIQRR353 pKa = 11.84GFEE356 pKa = 4.03ADD358 pKa = 3.86GDD360 pKa = 4.09LSACNTWTADD370 pKa = 3.64LEE372 pKa = 4.56QVTT375 pKa = 4.3
Molecular weight: 40.17 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C7MBM7|C7MBM7_BRAFD Predicted aminoglycoside phosphotransferase OS=Brachybacterium faecium (strain ATCC 43885 / DSM 4810 / JCM 11609 / LMG 19847 / NBRC 14762 / NCIMB 9860 / 6-10) OX=446465 GN=Bfae_11380 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 8.99RR4 pKa = 11.84TFQPNLRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 9.98KK16 pKa = 9.08HH17 pKa = 4.39GFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILNARR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.29GRR40 pKa = 11.84SEE42 pKa = 4.21LSAA45 pKa = 4.73
MM1 pKa = 7.69SKK3 pKa = 8.99RR4 pKa = 11.84TFQPNLRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 9.98KK16 pKa = 9.08HH17 pKa = 4.39GFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILNARR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.29GRR40 pKa = 11.84SEE42 pKa = 4.21LSAA45 pKa = 4.73
Molecular weight: 5.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1070770 |
33 |
2084 |
349.7 |
37.43 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.407 ± 0.068 | 0.532 ± 0.008 |
6.055 ± 0.041 | 6.583 ± 0.047 |
2.757 ± 0.028 | 9.146 ± 0.038 |
2.305 ± 0.02 | 3.932 ± 0.031 |
1.556 ± 0.029 | 10.758 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.945 ± 0.016 | 1.573 ± 0.022 |
5.89 ± 0.037 | 2.958 ± 0.021 |
7.64 ± 0.048 | 5.522 ± 0.031 |
6.004 ± 0.028 | 8.224 ± 0.041 |
1.432 ± 0.017 | 1.784 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
