
Avon-Heathcote Estuary associated circular virus 28
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; unclassified Circoviridae
Average proteome isoelectric point is 7.58
Get precalculated fractions of proteins
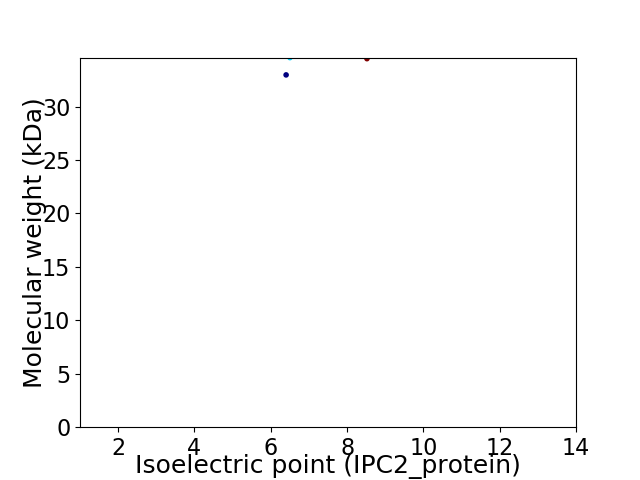
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
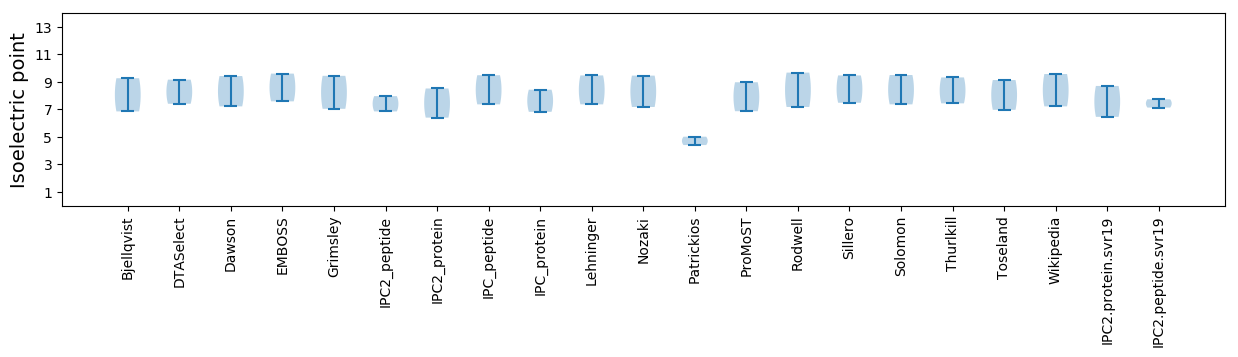
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0C5IMN2|A0A0C5IMN2_9CIRC Putative capsid protein OS=Avon-Heathcote Estuary associated circular virus 28 OX=1618252 PE=4 SV=1
MM1 pKa = 7.52ARR3 pKa = 11.84THH5 pKa = 6.42AKK7 pKa = 10.35KK8 pKa = 8.92NTSKK12 pKa = 11.0DD13 pKa = 3.18PSFRR17 pKa = 11.84KK18 pKa = 8.05TKK20 pKa = 9.43PLEE23 pKa = 3.86YY24 pKa = 10.54LPVQRR29 pKa = 11.84KK30 pKa = 6.77ITLGTVSGATTYY42 pKa = 11.63GLIDD46 pKa = 3.92AGRR49 pKa = 11.84LLSTSNHH56 pKa = 5.08RR57 pKa = 11.84LYY59 pKa = 10.63RR60 pKa = 11.84YY61 pKa = 8.81GKK63 pKa = 10.03RR64 pKa = 11.84YY65 pKa = 8.92NLKK68 pKa = 9.73IDD70 pKa = 3.64IDD72 pKa = 4.04PSVAATNTIEE82 pKa = 3.88VWALADD88 pKa = 3.21TWAVQKK94 pKa = 10.95AFEE97 pKa = 3.92EE98 pKa = 4.33AKK100 pKa = 10.44RR101 pKa = 11.84VFDD104 pKa = 3.95KK105 pKa = 11.15AYY107 pKa = 8.58TNEE110 pKa = 4.05RR111 pKa = 11.84EE112 pKa = 4.21NLSKK116 pKa = 10.71DD117 pKa = 3.01ARR119 pKa = 11.84ARR121 pKa = 11.84WFDD124 pKa = 3.36FRR126 pKa = 11.84ASIGVSGDD134 pKa = 2.46IMMAAVSTDD143 pKa = 3.89PNPGTLTLLTGGEE156 pKa = 4.2FANSIVEE163 pKa = 4.3DD164 pKa = 3.58NAGVQRR170 pKa = 11.84AFSWSPATSASTYY183 pKa = 10.16SIPAEE188 pKa = 3.8YY189 pKa = 10.56DD190 pKa = 3.22LAGNTSTSPTTPTGAGPYY208 pKa = 10.15DD209 pKa = 4.26DD210 pKa = 5.57LQADD214 pKa = 3.99SSAIEE219 pKa = 4.06MEE221 pKa = 4.11ALQDD225 pKa = 3.7RR226 pKa = 11.84GNLPPYY232 pKa = 9.0EE233 pKa = 4.92ANTFPPVWVKK243 pKa = 10.43IATLGQSAAGSQRR256 pKa = 11.84LSTGFFDD263 pKa = 4.53APCGQVVLSSSAPITEE279 pKa = 4.45LTGKK283 pKa = 10.34AYY285 pKa = 10.42AEE287 pKa = 4.21FKK289 pKa = 10.54IGDD292 pKa = 3.76YY293 pKa = 10.81KK294 pKa = 10.88GVKK297 pKa = 8.97AHH299 pKa = 6.04NLEE302 pKa = 4.26RR303 pKa = 11.84MM304 pKa = 3.65
MM1 pKa = 7.52ARR3 pKa = 11.84THH5 pKa = 6.42AKK7 pKa = 10.35KK8 pKa = 8.92NTSKK12 pKa = 11.0DD13 pKa = 3.18PSFRR17 pKa = 11.84KK18 pKa = 8.05TKK20 pKa = 9.43PLEE23 pKa = 3.86YY24 pKa = 10.54LPVQRR29 pKa = 11.84KK30 pKa = 6.77ITLGTVSGATTYY42 pKa = 11.63GLIDD46 pKa = 3.92AGRR49 pKa = 11.84LLSTSNHH56 pKa = 5.08RR57 pKa = 11.84LYY59 pKa = 10.63RR60 pKa = 11.84YY61 pKa = 8.81GKK63 pKa = 10.03RR64 pKa = 11.84YY65 pKa = 8.92NLKK68 pKa = 9.73IDD70 pKa = 3.64IDD72 pKa = 4.04PSVAATNTIEE82 pKa = 3.88VWALADD88 pKa = 3.21TWAVQKK94 pKa = 10.95AFEE97 pKa = 3.92EE98 pKa = 4.33AKK100 pKa = 10.44RR101 pKa = 11.84VFDD104 pKa = 3.95KK105 pKa = 11.15AYY107 pKa = 8.58TNEE110 pKa = 4.05RR111 pKa = 11.84EE112 pKa = 4.21NLSKK116 pKa = 10.71DD117 pKa = 3.01ARR119 pKa = 11.84ARR121 pKa = 11.84WFDD124 pKa = 3.36FRR126 pKa = 11.84ASIGVSGDD134 pKa = 2.46IMMAAVSTDD143 pKa = 3.89PNPGTLTLLTGGEE156 pKa = 4.2FANSIVEE163 pKa = 4.3DD164 pKa = 3.58NAGVQRR170 pKa = 11.84AFSWSPATSASTYY183 pKa = 10.16SIPAEE188 pKa = 3.8YY189 pKa = 10.56DD190 pKa = 3.22LAGNTSTSPTTPTGAGPYY208 pKa = 10.15DD209 pKa = 4.26DD210 pKa = 5.57LQADD214 pKa = 3.99SSAIEE219 pKa = 4.06MEE221 pKa = 4.11ALQDD225 pKa = 3.7RR226 pKa = 11.84GNLPPYY232 pKa = 9.0EE233 pKa = 4.92ANTFPPVWVKK243 pKa = 10.43IATLGQSAAGSQRR256 pKa = 11.84LSTGFFDD263 pKa = 4.53APCGQVVLSSSAPITEE279 pKa = 4.45LTGKK283 pKa = 10.34AYY285 pKa = 10.42AEE287 pKa = 4.21FKK289 pKa = 10.54IGDD292 pKa = 3.76YY293 pKa = 10.81KK294 pKa = 10.88GVKK297 pKa = 8.97AHH299 pKa = 6.04NLEE302 pKa = 4.26RR303 pKa = 11.84MM304 pKa = 3.65
Molecular weight: 32.97 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0C5IMN2|A0A0C5IMN2_9CIRC Putative capsid protein OS=Avon-Heathcote Estuary associated circular virus 28 OX=1618252 PE=4 SV=1
MM1 pKa = 6.99LQEE4 pKa = 4.65SIVRR8 pKa = 11.84TPRR11 pKa = 11.84FPPNVCWGVSGKK23 pKa = 9.44WEE25 pKa = 4.1DD26 pKa = 3.29QCMKK30 pKa = 10.66VEE32 pKa = 4.06ATPDD36 pKa = 3.0GCLNARR42 pKa = 11.84QFLNDD47 pKa = 3.48RR48 pKa = 11.84SLCPRR53 pKa = 11.84WRR55 pKa = 11.84LCPGCEE61 pKa = 3.58KK62 pKa = 10.51VRR64 pKa = 11.84AKK66 pKa = 10.55RR67 pKa = 11.84NQYY70 pKa = 10.39KK71 pKa = 9.4IAKK74 pKa = 9.03RR75 pKa = 11.84LEE77 pKa = 4.2FDD79 pKa = 4.64LEE81 pKa = 4.12WAKK84 pKa = 10.78EE85 pKa = 3.54ADD87 pKa = 3.4IPLKK91 pKa = 10.97VGVLTTTLPGKK102 pKa = 9.81EE103 pKa = 3.59SWIRR107 pKa = 11.84NASLGEE113 pKa = 3.93QYY115 pKa = 11.19SYY117 pKa = 10.12LTEE120 pKa = 3.97RR121 pKa = 11.84RR122 pKa = 11.84TMSGYY127 pKa = 9.08TGWHH131 pKa = 5.56SMRR134 pKa = 11.84GLNTKK139 pKa = 10.09LKK141 pKa = 9.52EE142 pKa = 3.84WGISGGSHH150 pKa = 5.16YY151 pKa = 11.26LEE153 pKa = 4.47FTNKK157 pKa = 7.65GTTWNTHH164 pKa = 3.61MHH166 pKa = 6.49SILVGFQDD174 pKa = 4.04DD175 pKa = 3.71WQVPLKK181 pKa = 9.47EE182 pKa = 4.34TTKK185 pKa = 10.33QLEE188 pKa = 4.41WNDD191 pKa = 3.92DD192 pKa = 3.1LTMRR196 pKa = 11.84LQTEE200 pKa = 3.82KK201 pKa = 11.24AEE203 pKa = 4.15NKK205 pKa = 8.3TRR207 pKa = 11.84SNKK210 pKa = 10.14RR211 pKa = 11.84ILEE214 pKa = 3.92PLGLGRR220 pKa = 11.84LYY222 pKa = 10.6TLDD225 pKa = 3.36IASDD229 pKa = 4.05DD230 pKa = 3.91EE231 pKa = 4.3LASIARR237 pKa = 11.84YY238 pKa = 7.54SAKK241 pKa = 10.16VEE243 pKa = 4.26YY244 pKa = 8.82VTKK247 pKa = 9.96PVKK250 pKa = 10.62VPEE253 pKa = 4.55GKK255 pKa = 10.5LVDD258 pKa = 3.74VTNFLAGGFEE268 pKa = 4.23HH269 pKa = 6.79GKK271 pKa = 9.37EE272 pKa = 3.88RR273 pKa = 11.84SGHH276 pKa = 5.01GRR278 pKa = 11.84HH279 pKa = 6.05LPRR282 pKa = 11.84LARR285 pKa = 11.84PFGDD289 pKa = 2.81WMKK292 pKa = 10.82NGPEE296 pKa = 3.43RR297 pKa = 11.84RR298 pKa = 11.84YY299 pKa = 10.62AA300 pKa = 3.48
MM1 pKa = 6.99LQEE4 pKa = 4.65SIVRR8 pKa = 11.84TPRR11 pKa = 11.84FPPNVCWGVSGKK23 pKa = 9.44WEE25 pKa = 4.1DD26 pKa = 3.29QCMKK30 pKa = 10.66VEE32 pKa = 4.06ATPDD36 pKa = 3.0GCLNARR42 pKa = 11.84QFLNDD47 pKa = 3.48RR48 pKa = 11.84SLCPRR53 pKa = 11.84WRR55 pKa = 11.84LCPGCEE61 pKa = 3.58KK62 pKa = 10.51VRR64 pKa = 11.84AKK66 pKa = 10.55RR67 pKa = 11.84NQYY70 pKa = 10.39KK71 pKa = 9.4IAKK74 pKa = 9.03RR75 pKa = 11.84LEE77 pKa = 4.2FDD79 pKa = 4.64LEE81 pKa = 4.12WAKK84 pKa = 10.78EE85 pKa = 3.54ADD87 pKa = 3.4IPLKK91 pKa = 10.97VGVLTTTLPGKK102 pKa = 9.81EE103 pKa = 3.59SWIRR107 pKa = 11.84NASLGEE113 pKa = 3.93QYY115 pKa = 11.19SYY117 pKa = 10.12LTEE120 pKa = 3.97RR121 pKa = 11.84RR122 pKa = 11.84TMSGYY127 pKa = 9.08TGWHH131 pKa = 5.56SMRR134 pKa = 11.84GLNTKK139 pKa = 10.09LKK141 pKa = 9.52EE142 pKa = 3.84WGISGGSHH150 pKa = 5.16YY151 pKa = 11.26LEE153 pKa = 4.47FTNKK157 pKa = 7.65GTTWNTHH164 pKa = 3.61MHH166 pKa = 6.49SILVGFQDD174 pKa = 4.04DD175 pKa = 3.71WQVPLKK181 pKa = 9.47EE182 pKa = 4.34TTKK185 pKa = 10.33QLEE188 pKa = 4.41WNDD191 pKa = 3.92DD192 pKa = 3.1LTMRR196 pKa = 11.84LQTEE200 pKa = 3.82KK201 pKa = 11.24AEE203 pKa = 4.15NKK205 pKa = 8.3TRR207 pKa = 11.84SNKK210 pKa = 10.14RR211 pKa = 11.84ILEE214 pKa = 3.92PLGLGRR220 pKa = 11.84LYY222 pKa = 10.6TLDD225 pKa = 3.36IASDD229 pKa = 4.05DD230 pKa = 3.91EE231 pKa = 4.3LASIARR237 pKa = 11.84YY238 pKa = 7.54SAKK241 pKa = 10.16VEE243 pKa = 4.26YY244 pKa = 8.82VTKK247 pKa = 9.96PVKK250 pKa = 10.62VPEE253 pKa = 4.55GKK255 pKa = 10.5LVDD258 pKa = 3.74VTNFLAGGFEE268 pKa = 4.23HH269 pKa = 6.79GKK271 pKa = 9.37EE272 pKa = 3.88RR273 pKa = 11.84SGHH276 pKa = 5.01GRR278 pKa = 11.84HH279 pKa = 6.05LPRR282 pKa = 11.84LARR285 pKa = 11.84PFGDD289 pKa = 2.81WMKK292 pKa = 10.82NGPEE296 pKa = 3.43RR297 pKa = 11.84RR298 pKa = 11.84YY299 pKa = 10.62AA300 pKa = 3.48
Molecular weight: 34.49 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
604 |
300 |
304 |
302.0 |
33.73 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.609 ± 2.535 | 1.159 ± 0.591 |
5.298 ± 0.443 | 6.291 ± 0.966 |
3.146 ± 0.336 | 7.781 ± 0.388 |
1.656 ± 0.476 | 3.642 ± 0.451 |
6.623 ± 0.733 | 8.609 ± 0.977 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.987 ± 0.243 | 4.305 ± 0.02 |
5.298 ± 0.209 | 2.815 ± 0.13 |
6.623 ± 0.968 | 6.954 ± 1.138 |
8.113 ± 0.782 | 4.967 ± 0.023 |
2.649 ± 0.715 | 3.477 ± 0.335 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
