
Rice stripe necrosis virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Hepelivirales; Benyviridae; Benyvirus
Average proteome isoelectric point is 6.49
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|B5STA0|B5STA0_9VIRU 12K protein OS=Rice stripe necrosis virus OX=373373 GN=TGBp2 PE=4 SV=1
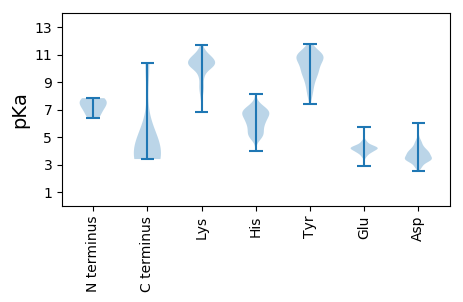
MM1 pKa = 6.36TSEE4 pKa = 4.24WARR7 pKa = 11.84EE8 pKa = 3.87HH9 pKa = 6.1PTDD12 pKa = 3.23VFAIFEE18 pKa = 4.32TCARR22 pKa = 11.84EE23 pKa = 5.74AGFTWNDD30 pKa = 3.35VPPHH34 pKa = 5.41VVNFDD39 pKa = 3.57SLEE42 pKa = 3.92KK43 pKa = 10.78SKK45 pKa = 11.05AISNLTDD52 pKa = 3.66LLEE55 pKa = 4.68NEE57 pKa = 4.36VAKK60 pKa = 10.78GCKK63 pKa = 9.46EE64 pKa = 4.09KK65 pKa = 11.28GDD67 pKa = 3.51VAAIKK72 pKa = 10.55LEE74 pKa = 4.1NVNGMTKK81 pKa = 10.07EE82 pKa = 3.69YY83 pKa = 9.19NARR86 pKa = 11.84VGVCVGAPGAGKK98 pKa = 6.85TTLIKK103 pKa = 10.74AVMSKK108 pKa = 10.73ASRR111 pKa = 11.84VVIAVPNSTLLKK123 pKa = 10.21NVYY126 pKa = 9.84SGNPNAFLIDD136 pKa = 4.0DD137 pKa = 4.58LFSRR141 pKa = 11.84PVEE144 pKa = 3.78FAKK147 pKa = 11.17YY148 pKa = 7.38EE149 pKa = 4.16TILIDD154 pKa = 3.79EE155 pKa = 4.47FTKK158 pKa = 10.54VHH160 pKa = 4.94VCEE163 pKa = 4.07VLMLSALLRR172 pKa = 11.84VKK174 pKa = 10.51NILMFGDD181 pKa = 4.33PQQGMHH187 pKa = 6.0YY188 pKa = 9.81RR189 pKa = 11.84PGSFVYY195 pKa = 10.97YY196 pKa = 9.8NFPVLAEE203 pKa = 4.01SHH205 pKa = 6.52ASHH208 pKa = 7.1RR209 pKa = 11.84MPQAIGEE216 pKa = 4.36AYY218 pKa = 9.83NNAMGTKK225 pKa = 9.21IEE227 pKa = 4.48PKK229 pKa = 9.86SSQSGEE235 pKa = 4.1FEE237 pKa = 4.15IKK239 pKa = 10.47DD240 pKa = 3.97LLGMIRR246 pKa = 11.84DD247 pKa = 3.83KK248 pKa = 11.64SKK250 pKa = 10.92VLCLSEE256 pKa = 4.19KK257 pKa = 9.2TQSNLDD263 pKa = 3.5DD264 pKa = 4.77CGISAEE270 pKa = 4.54LVSKK274 pKa = 10.66VQGCEE279 pKa = 3.67FAAVTLILEE288 pKa = 4.65EE289 pKa = 4.48PQDD292 pKa = 3.63IAPFCNKK299 pKa = 10.25SIRR302 pKa = 11.84CVALSRR308 pKa = 11.84AKK310 pKa = 10.18EE311 pKa = 3.9VLIIQATPYY320 pKa = 9.35FKK322 pKa = 11.33SMLCNAEE329 pKa = 4.13FVDD332 pKa = 4.74PYY334 pKa = 10.87EE335 pKa = 4.4VDD337 pKa = 3.74SSCSGQTLCNSRR349 pKa = 3.74
MM1 pKa = 6.36TSEE4 pKa = 4.24WARR7 pKa = 11.84EE8 pKa = 3.87HH9 pKa = 6.1PTDD12 pKa = 3.23VFAIFEE18 pKa = 4.32TCARR22 pKa = 11.84EE23 pKa = 5.74AGFTWNDD30 pKa = 3.35VPPHH34 pKa = 5.41VVNFDD39 pKa = 3.57SLEE42 pKa = 3.92KK43 pKa = 10.78SKK45 pKa = 11.05AISNLTDD52 pKa = 3.66LLEE55 pKa = 4.68NEE57 pKa = 4.36VAKK60 pKa = 10.78GCKK63 pKa = 9.46EE64 pKa = 4.09KK65 pKa = 11.28GDD67 pKa = 3.51VAAIKK72 pKa = 10.55LEE74 pKa = 4.1NVNGMTKK81 pKa = 10.07EE82 pKa = 3.69YY83 pKa = 9.19NARR86 pKa = 11.84VGVCVGAPGAGKK98 pKa = 6.85TTLIKK103 pKa = 10.74AVMSKK108 pKa = 10.73ASRR111 pKa = 11.84VVIAVPNSTLLKK123 pKa = 10.21NVYY126 pKa = 9.84SGNPNAFLIDD136 pKa = 4.0DD137 pKa = 4.58LFSRR141 pKa = 11.84PVEE144 pKa = 3.78FAKK147 pKa = 11.17YY148 pKa = 7.38EE149 pKa = 4.16TILIDD154 pKa = 3.79EE155 pKa = 4.47FTKK158 pKa = 10.54VHH160 pKa = 4.94VCEE163 pKa = 4.07VLMLSALLRR172 pKa = 11.84VKK174 pKa = 10.51NILMFGDD181 pKa = 4.33PQQGMHH187 pKa = 6.0YY188 pKa = 9.81RR189 pKa = 11.84PGSFVYY195 pKa = 10.97YY196 pKa = 9.8NFPVLAEE203 pKa = 4.01SHH205 pKa = 6.52ASHH208 pKa = 7.1RR209 pKa = 11.84MPQAIGEE216 pKa = 4.36AYY218 pKa = 9.83NNAMGTKK225 pKa = 9.21IEE227 pKa = 4.48PKK229 pKa = 9.86SSQSGEE235 pKa = 4.1FEE237 pKa = 4.15IKK239 pKa = 10.47DD240 pKa = 3.97LLGMIRR246 pKa = 11.84DD247 pKa = 3.83KK248 pKa = 11.64SKK250 pKa = 10.92VLCLSEE256 pKa = 4.19KK257 pKa = 9.2TQSNLDD263 pKa = 3.5DD264 pKa = 4.77CGISAEE270 pKa = 4.54LVSKK274 pKa = 10.66VQGCEE279 pKa = 3.67FAAVTLILEE288 pKa = 4.65EE289 pKa = 4.48PQDD292 pKa = 3.63IAPFCNKK299 pKa = 10.25SIRR302 pKa = 11.84CVALSRR308 pKa = 11.84AKK310 pKa = 10.18EE311 pKa = 3.9VLIIQATPYY320 pKa = 9.35FKK322 pKa = 11.33SMLCNAEE329 pKa = 4.13FVDD332 pKa = 4.74PYY334 pKa = 10.87EE335 pKa = 4.4VDD337 pKa = 3.74SSCSGQTLCNSRR349 pKa = 3.74
Molecular weight: 38.42 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|B5STA1|B5STA1_9VIRU 15K protein OS=Rice stripe necrosis virus OX=373373 GN=TGBp3 PE=4 SV=2
MM1 pKa = 7.35SGQYY5 pKa = 10.44VSARR9 pKa = 11.84PNKK12 pKa = 9.89FIPICICIGVVCVAVCLVLATPRR35 pKa = 11.84HH36 pKa = 5.19KK37 pKa = 9.4THH39 pKa = 6.51SAGDD43 pKa = 3.61YY44 pKa = 9.83GVPTFANGGSYY55 pKa = 11.33ADD57 pKa = 3.45GTRR60 pKa = 11.84RR61 pKa = 11.84AKK63 pKa = 10.46FNCNNDD69 pKa = 3.2RR70 pKa = 11.84AYY72 pKa = 10.64GSSQPQMSSNFVAFIAVILLVAFALRR98 pKa = 11.84SCNSVGNCGEE108 pKa = 4.11RR109 pKa = 11.84CNGGCCKK116 pKa = 10.55NN117 pKa = 3.46
MM1 pKa = 7.35SGQYY5 pKa = 10.44VSARR9 pKa = 11.84PNKK12 pKa = 9.89FIPICICIGVVCVAVCLVLATPRR35 pKa = 11.84HH36 pKa = 5.19KK37 pKa = 9.4THH39 pKa = 6.51SAGDD43 pKa = 3.61YY44 pKa = 9.83GVPTFANGGSYY55 pKa = 11.33ADD57 pKa = 3.45GTRR60 pKa = 11.84RR61 pKa = 11.84AKK63 pKa = 10.46FNCNNDD69 pKa = 3.2RR70 pKa = 11.84AYY72 pKa = 10.64GSSQPQMSSNFVAFIAVILLVAFALRR98 pKa = 11.84SCNSVGNCGEE108 pKa = 4.11RR109 pKa = 11.84CNGGCCKK116 pKa = 10.55NN117 pKa = 3.46
Molecular weight: 12.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4177 |
117 |
2100 |
522.1 |
58.27 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.283 ± 0.225 | 1.819 ± 0.648 |
5.602 ± 0.276 | 6.416 ± 0.581 |
4.166 ± 0.302 | 6.584 ± 0.538 |
1.891 ± 0.202 | 4.956 ± 0.306 |
4.812 ± 0.534 | 8.188 ± 0.432 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.657 ± 0.293 | 4.597 ± 0.282 |
4.98 ± 0.447 | 3.663 ± 0.41 |
6.512 ± 0.743 | 7.709 ± 0.679 |
4.74 ± 0.237 | 7.541 ± 0.831 |
1.269 ± 0.241 | 3.543 ± 0.258 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
