
Hubei sobemo-like virus 42
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.84
Get precalculated fractions of proteins
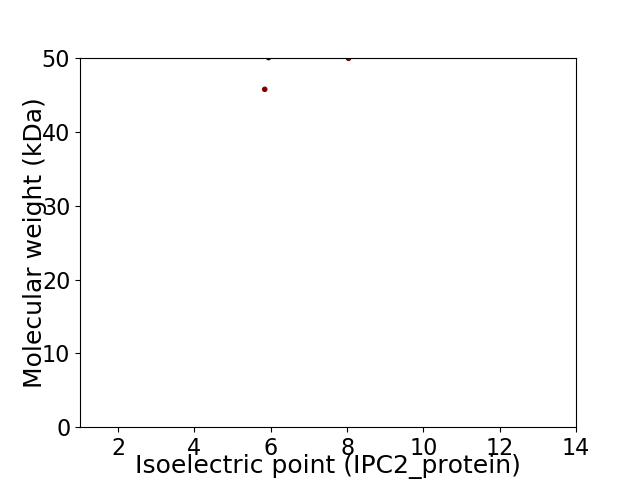
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A1L3KEL0|A0A1L3KEL0_9VIRU Uncharacterized protein OS=Hubei sobemo-like virus 42 OX=1923230 PE=4 SV=1
MM1 pKa = 7.96DD2 pKa = 5.97RR3 pKa = 11.84ILYY6 pKa = 10.39VMEE9 pKa = 3.79QLYY12 pKa = 10.34EE13 pKa = 4.21SVRR16 pKa = 11.84VDD18 pKa = 3.21HH19 pKa = 6.69YY20 pKa = 12.09NWFCYY25 pKa = 10.74DD26 pKa = 3.15NFLKK30 pKa = 10.69VVRR33 pKa = 11.84CLEE36 pKa = 4.34FSSSPGYY43 pKa = 10.49GFSRR47 pKa = 11.84TAPTIGGWLKK57 pKa = 10.56FDD59 pKa = 4.53GMNFDD64 pKa = 3.94PQRR67 pKa = 11.84LSEE70 pKa = 4.3LWHH73 pKa = 5.87MVNVCYY79 pKa = 10.4RR80 pKa = 11.84EE81 pKa = 4.18PKK83 pKa = 10.15LFSYY87 pKa = 9.59WKK89 pKa = 10.08VFIKK93 pKa = 10.57RR94 pKa = 11.84EE95 pKa = 3.84PHH97 pKa = 5.54KK98 pKa = 9.61TSKK101 pKa = 10.09MEE103 pKa = 3.96NKK105 pKa = 9.24RR106 pKa = 11.84WRR108 pKa = 11.84LIQCCPLDD116 pKa = 4.22VQVLWHH122 pKa = 5.95MLFAKK127 pKa = 10.59SNEE130 pKa = 3.99LEE132 pKa = 4.4ANEE135 pKa = 4.37SMSIPSVQGMVLPFGGWKK153 pKa = 8.62EE154 pKa = 4.14HH155 pKa = 5.37YY156 pKa = 9.68NRR158 pKa = 11.84WMKK161 pKa = 10.99NKK163 pKa = 10.11LFFGSDD169 pKa = 2.56KK170 pKa = 10.88TAWDD174 pKa = 3.45WTASEE179 pKa = 4.16WMIEE183 pKa = 3.77LDD185 pKa = 3.36LEE187 pKa = 4.08LRR189 pKa = 11.84RR190 pKa = 11.84RR191 pKa = 11.84LIHH194 pKa = 7.15TDD196 pKa = 3.69DD197 pKa = 3.86DD198 pKa = 4.95WITQAAKK205 pKa = 9.94VYY207 pKa = 10.67EE208 pKa = 4.07NAFYY212 pKa = 10.48DD213 pKa = 3.33AHH215 pKa = 7.15LILGDD220 pKa = 3.41GRR222 pKa = 11.84IYY224 pKa = 10.44RR225 pKa = 11.84QMYY228 pKa = 8.91PGIIKK233 pKa = 10.05SGCVNTISINSRR245 pKa = 11.84CQVMLHH251 pKa = 6.52LLYY254 pKa = 10.7SFRR257 pKa = 11.84KK258 pKa = 9.7GISPYY263 pKa = 10.33PMVVAVGDD271 pKa = 3.76DD272 pKa = 3.43TLQAEE277 pKa = 4.41EE278 pKa = 4.44HH279 pKa = 6.64AVDD282 pKa = 3.12KK283 pKa = 11.21QMYY286 pKa = 8.45EE287 pKa = 3.81RR288 pKa = 11.84FGVVIKK294 pKa = 10.76SVSEE298 pKa = 3.77TLEE301 pKa = 3.72FLGRR305 pKa = 11.84EE306 pKa = 4.25WNDD309 pKa = 3.27DD310 pKa = 3.79GPRR313 pKa = 11.84PMYY316 pKa = 9.88VSKK319 pKa = 10.7HH320 pKa = 5.19IFSLCYY326 pKa = 10.28KK327 pKa = 10.11NIEE330 pKa = 4.27LTPQILDD337 pKa = 3.23ALMRR341 pKa = 11.84EE342 pKa = 4.49YY343 pKa = 11.42VNEE346 pKa = 3.78DD347 pKa = 3.19SMFNFLTILTRR358 pKa = 11.84EE359 pKa = 4.36LGFSSNVHH367 pKa = 5.3SRR369 pKa = 11.84DD370 pKa = 3.5YY371 pKa = 10.79YY372 pKa = 10.75KK373 pKa = 11.14YY374 pKa = 10.57WMDD377 pKa = 4.56NPDD380 pKa = 3.48AQYY383 pKa = 11.69LHH385 pKa = 7.0
MM1 pKa = 7.96DD2 pKa = 5.97RR3 pKa = 11.84ILYY6 pKa = 10.39VMEE9 pKa = 3.79QLYY12 pKa = 10.34EE13 pKa = 4.21SVRR16 pKa = 11.84VDD18 pKa = 3.21HH19 pKa = 6.69YY20 pKa = 12.09NWFCYY25 pKa = 10.74DD26 pKa = 3.15NFLKK30 pKa = 10.69VVRR33 pKa = 11.84CLEE36 pKa = 4.34FSSSPGYY43 pKa = 10.49GFSRR47 pKa = 11.84TAPTIGGWLKK57 pKa = 10.56FDD59 pKa = 4.53GMNFDD64 pKa = 3.94PQRR67 pKa = 11.84LSEE70 pKa = 4.3LWHH73 pKa = 5.87MVNVCYY79 pKa = 10.4RR80 pKa = 11.84EE81 pKa = 4.18PKK83 pKa = 10.15LFSYY87 pKa = 9.59WKK89 pKa = 10.08VFIKK93 pKa = 10.57RR94 pKa = 11.84EE95 pKa = 3.84PHH97 pKa = 5.54KK98 pKa = 9.61TSKK101 pKa = 10.09MEE103 pKa = 3.96NKK105 pKa = 9.24RR106 pKa = 11.84WRR108 pKa = 11.84LIQCCPLDD116 pKa = 4.22VQVLWHH122 pKa = 5.95MLFAKK127 pKa = 10.59SNEE130 pKa = 3.99LEE132 pKa = 4.4ANEE135 pKa = 4.37SMSIPSVQGMVLPFGGWKK153 pKa = 8.62EE154 pKa = 4.14HH155 pKa = 5.37YY156 pKa = 9.68NRR158 pKa = 11.84WMKK161 pKa = 10.99NKK163 pKa = 10.11LFFGSDD169 pKa = 2.56KK170 pKa = 10.88TAWDD174 pKa = 3.45WTASEE179 pKa = 4.16WMIEE183 pKa = 3.77LDD185 pKa = 3.36LEE187 pKa = 4.08LRR189 pKa = 11.84RR190 pKa = 11.84RR191 pKa = 11.84LIHH194 pKa = 7.15TDD196 pKa = 3.69DD197 pKa = 3.86DD198 pKa = 4.95WITQAAKK205 pKa = 9.94VYY207 pKa = 10.67EE208 pKa = 4.07NAFYY212 pKa = 10.48DD213 pKa = 3.33AHH215 pKa = 7.15LILGDD220 pKa = 3.41GRR222 pKa = 11.84IYY224 pKa = 10.44RR225 pKa = 11.84QMYY228 pKa = 8.91PGIIKK233 pKa = 10.05SGCVNTISINSRR245 pKa = 11.84CQVMLHH251 pKa = 6.52LLYY254 pKa = 10.7SFRR257 pKa = 11.84KK258 pKa = 9.7GISPYY263 pKa = 10.33PMVVAVGDD271 pKa = 3.76DD272 pKa = 3.43TLQAEE277 pKa = 4.41EE278 pKa = 4.44HH279 pKa = 6.64AVDD282 pKa = 3.12KK283 pKa = 11.21QMYY286 pKa = 8.45EE287 pKa = 3.81RR288 pKa = 11.84FGVVIKK294 pKa = 10.76SVSEE298 pKa = 3.77TLEE301 pKa = 3.72FLGRR305 pKa = 11.84EE306 pKa = 4.25WNDD309 pKa = 3.27DD310 pKa = 3.79GPRR313 pKa = 11.84PMYY316 pKa = 9.88VSKK319 pKa = 10.7HH320 pKa = 5.19IFSLCYY326 pKa = 10.28KK327 pKa = 10.11NIEE330 pKa = 4.27LTPQILDD337 pKa = 3.23ALMRR341 pKa = 11.84EE342 pKa = 4.49YY343 pKa = 11.42VNEE346 pKa = 3.78DD347 pKa = 3.19SMFNFLTILTRR358 pKa = 11.84EE359 pKa = 4.36LGFSSNVHH367 pKa = 5.3SRR369 pKa = 11.84DD370 pKa = 3.5YY371 pKa = 10.79YY372 pKa = 10.75KK373 pKa = 11.14YY374 pKa = 10.57WMDD377 pKa = 4.56NPDD380 pKa = 3.48AQYY383 pKa = 11.69LHH385 pKa = 7.0
Molecular weight: 45.76 kDa
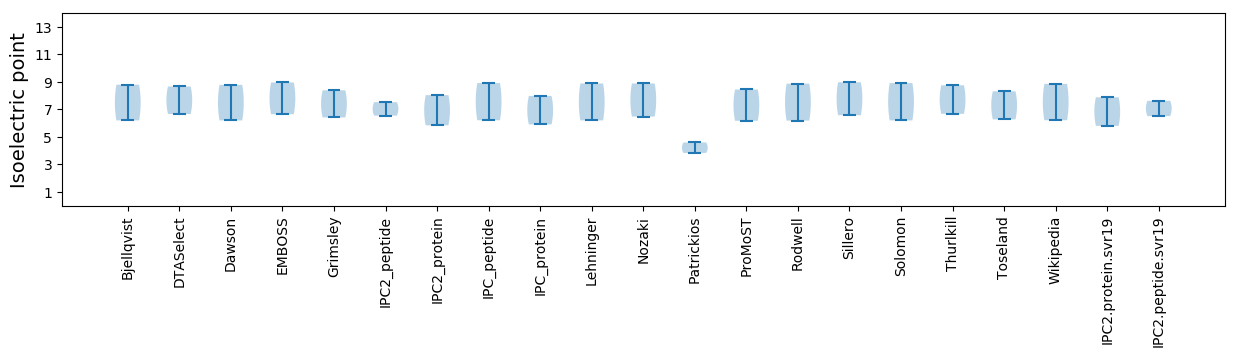
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KEL0|A0A1L3KEL0_9VIRU Uncharacterized protein OS=Hubei sobemo-like virus 42 OX=1923230 PE=4 SV=1
MM1 pKa = 7.9EE2 pKa = 5.62DD3 pKa = 3.29QTVLALIKK11 pKa = 10.52QLVRR15 pKa = 11.84TMGSNAWNLFKK26 pKa = 11.13DD27 pKa = 4.52FLTSSYY33 pKa = 10.16VRR35 pKa = 11.84NFAALAVSSMALYY48 pKa = 10.73SRR50 pKa = 11.84INSWLRR56 pKa = 11.84EE57 pKa = 4.08NNTSILEE64 pKa = 4.06YY65 pKa = 10.73LLKK68 pKa = 10.0TSKK71 pKa = 10.82LFLSVTKK78 pKa = 10.3ILSWFRR84 pKa = 11.84KK85 pKa = 9.09PKK87 pKa = 10.23DD88 pKa = 3.42PRR90 pKa = 11.84PMVRR94 pKa = 11.84TSWAAQRR101 pKa = 11.84FPEE104 pKa = 4.67SIVQGSEE111 pKa = 3.71EE112 pKa = 4.01TEE114 pKa = 4.02MFHH117 pKa = 7.48KK118 pKa = 10.42RR119 pKa = 11.84GEE121 pKa = 4.12ALVVDD126 pKa = 4.29VNGNAVGKK134 pKa = 9.39GARR137 pKa = 11.84FWNVLVVPEE146 pKa = 4.54HH147 pKa = 6.65VIGTCRR153 pKa = 11.84YY154 pKa = 8.06SSGSSVVYY162 pKa = 9.66ICHH165 pKa = 7.63PDD167 pKa = 3.34RR168 pKa = 11.84PDD170 pKa = 3.91CKK172 pKa = 10.62FEE174 pKa = 4.11LAGEE178 pKa = 4.17PVFLYY183 pKa = 10.88SDD185 pKa = 3.9LLGYY189 pKa = 10.0NVSSSTWSQLGISQVSIGWITKK211 pKa = 7.29PTAASVVGVKK221 pKa = 10.37RR222 pKa = 11.84FGTSGILKK230 pKa = 10.11VDD232 pKa = 3.82DD233 pKa = 4.27EE234 pKa = 5.21SFGSVRR240 pKa = 11.84YY241 pKa = 7.99MATTCHH247 pKa = 6.67GYY249 pKa = 10.3SGALYY254 pKa = 9.95TSGNLTLGIHH264 pKa = 6.64LNGGKK269 pKa = 10.1KK270 pKa = 10.1NLGIAADD277 pKa = 4.17FVRR280 pKa = 11.84ALVSSKK286 pKa = 9.18MDD288 pKa = 3.11KK289 pKa = 10.76TMEE292 pKa = 4.45SSDD295 pKa = 3.71DD296 pKa = 3.83DD297 pKa = 3.4TWDD300 pKa = 4.89FINQRR305 pKa = 11.84WFDD308 pKa = 3.63GGKK311 pKa = 9.94LRR313 pKa = 11.84DD314 pKa = 3.96GVKK317 pKa = 10.09VDD319 pKa = 4.48YY320 pKa = 9.52ITADD324 pKa = 3.64EE325 pKa = 5.24IYY327 pKa = 10.38ASSQGRR333 pKa = 11.84FHH335 pKa = 7.14VVNKK339 pKa = 10.56DD340 pKa = 3.28VFRR343 pKa = 11.84KK344 pKa = 9.33HH345 pKa = 6.72ASEE348 pKa = 4.24EE349 pKa = 3.98QWNSLVYY356 pKa = 10.48ADD358 pKa = 5.17RR359 pKa = 11.84EE360 pKa = 4.43VEE362 pKa = 4.57SVFPLGAPRR371 pKa = 11.84LSPGASNMSEE381 pKa = 4.05KK382 pKa = 9.52VTAPTRR388 pKa = 11.84KK389 pKa = 9.68VISGGTAEE397 pKa = 5.2LQQQLQRR404 pKa = 11.84FAASPSGSRR413 pKa = 11.84SKK415 pKa = 10.93GAEE418 pKa = 3.66KK419 pKa = 10.16RR420 pKa = 11.84YY421 pKa = 9.46GWIRR425 pKa = 11.84ALTEE429 pKa = 4.21TEE431 pKa = 4.78LMDD434 pKa = 3.11YY435 pKa = 10.09WRR437 pKa = 11.84LLKK440 pKa = 10.28EE441 pKa = 4.19DD442 pKa = 4.33QEE444 pKa = 4.57NASQQ448 pKa = 3.65
MM1 pKa = 7.9EE2 pKa = 5.62DD3 pKa = 3.29QTVLALIKK11 pKa = 10.52QLVRR15 pKa = 11.84TMGSNAWNLFKK26 pKa = 11.13DD27 pKa = 4.52FLTSSYY33 pKa = 10.16VRR35 pKa = 11.84NFAALAVSSMALYY48 pKa = 10.73SRR50 pKa = 11.84INSWLRR56 pKa = 11.84EE57 pKa = 4.08NNTSILEE64 pKa = 4.06YY65 pKa = 10.73LLKK68 pKa = 10.0TSKK71 pKa = 10.82LFLSVTKK78 pKa = 10.3ILSWFRR84 pKa = 11.84KK85 pKa = 9.09PKK87 pKa = 10.23DD88 pKa = 3.42PRR90 pKa = 11.84PMVRR94 pKa = 11.84TSWAAQRR101 pKa = 11.84FPEE104 pKa = 4.67SIVQGSEE111 pKa = 3.71EE112 pKa = 4.01TEE114 pKa = 4.02MFHH117 pKa = 7.48KK118 pKa = 10.42RR119 pKa = 11.84GEE121 pKa = 4.12ALVVDD126 pKa = 4.29VNGNAVGKK134 pKa = 9.39GARR137 pKa = 11.84FWNVLVVPEE146 pKa = 4.54HH147 pKa = 6.65VIGTCRR153 pKa = 11.84YY154 pKa = 8.06SSGSSVVYY162 pKa = 9.66ICHH165 pKa = 7.63PDD167 pKa = 3.34RR168 pKa = 11.84PDD170 pKa = 3.91CKK172 pKa = 10.62FEE174 pKa = 4.11LAGEE178 pKa = 4.17PVFLYY183 pKa = 10.88SDD185 pKa = 3.9LLGYY189 pKa = 10.0NVSSSTWSQLGISQVSIGWITKK211 pKa = 7.29PTAASVVGVKK221 pKa = 10.37RR222 pKa = 11.84FGTSGILKK230 pKa = 10.11VDD232 pKa = 3.82DD233 pKa = 4.27EE234 pKa = 5.21SFGSVRR240 pKa = 11.84YY241 pKa = 7.99MATTCHH247 pKa = 6.67GYY249 pKa = 10.3SGALYY254 pKa = 9.95TSGNLTLGIHH264 pKa = 6.64LNGGKK269 pKa = 10.1KK270 pKa = 10.1NLGIAADD277 pKa = 4.17FVRR280 pKa = 11.84ALVSSKK286 pKa = 9.18MDD288 pKa = 3.11KK289 pKa = 10.76TMEE292 pKa = 4.45SSDD295 pKa = 3.71DD296 pKa = 3.83DD297 pKa = 3.4TWDD300 pKa = 4.89FINQRR305 pKa = 11.84WFDD308 pKa = 3.63GGKK311 pKa = 9.94LRR313 pKa = 11.84DD314 pKa = 3.96GVKK317 pKa = 10.09VDD319 pKa = 4.48YY320 pKa = 9.52ITADD324 pKa = 3.64EE325 pKa = 5.24IYY327 pKa = 10.38ASSQGRR333 pKa = 11.84FHH335 pKa = 7.14VVNKK339 pKa = 10.56DD340 pKa = 3.28VFRR343 pKa = 11.84KK344 pKa = 9.33HH345 pKa = 6.72ASEE348 pKa = 4.24EE349 pKa = 3.98QWNSLVYY356 pKa = 10.48ADD358 pKa = 5.17RR359 pKa = 11.84EE360 pKa = 4.43VEE362 pKa = 4.57SVFPLGAPRR371 pKa = 11.84LSPGASNMSEE381 pKa = 4.05KK382 pKa = 9.52VTAPTRR388 pKa = 11.84KK389 pKa = 9.68VISGGTAEE397 pKa = 5.2LQQQLQRR404 pKa = 11.84FAASPSGSRR413 pKa = 11.84SKK415 pKa = 10.93GAEE418 pKa = 3.66KK419 pKa = 10.16RR420 pKa = 11.84YY421 pKa = 9.46GWIRR425 pKa = 11.84ALTEE429 pKa = 4.21TEE431 pKa = 4.78LMDD434 pKa = 3.11YY435 pKa = 10.09WRR437 pKa = 11.84LLKK440 pKa = 10.28EE441 pKa = 4.19DD442 pKa = 4.33QEE444 pKa = 4.57NASQQ448 pKa = 3.65
Molecular weight: 49.95 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
833 |
385 |
448 |
416.5 |
47.86 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.522 ± 1.263 | 1.441 ± 0.427 |
5.642 ± 0.396 | 6.002 ± 0.329 |
4.682 ± 0.343 | 6.363 ± 0.956 |
2.281 ± 0.56 | 4.562 ± 0.424 |
5.642 ± 0.126 | 8.764 ± 0.219 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.361 ± 0.88 | 4.322 ± 0.237 |
3.481 ± 0.278 | 3.241 ± 0.083 |
6.002 ± 0.019 | 9.124 ± 1.413 |
4.562 ± 0.794 | 7.443 ± 0.636 |
3.121 ± 0.345 | 4.442 ± 0.852 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
