
Magnetospirillum gryphiswaldense (strain DSM 6361 / JCM 21280 / NBRC 15271 / MSR-1)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodospirillales; Rhodospirillaceae; Magnetospirillum; Magnetospirillum gryphiswaldense
Average proteome isoelectric point is 6.58
Get precalculated fractions of proteins
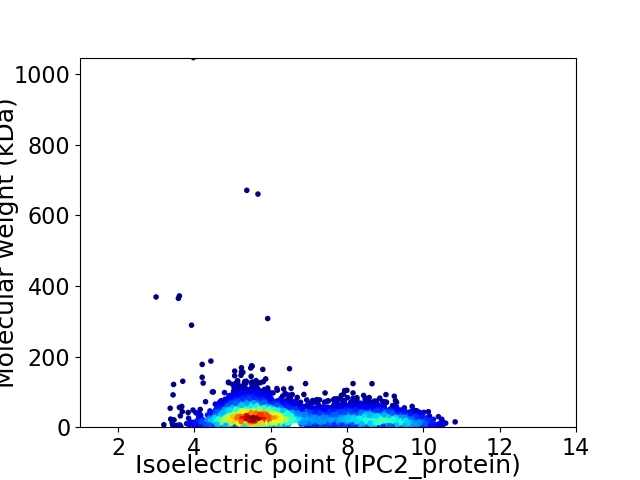
Virtual 2D-PAGE plot for 4155 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
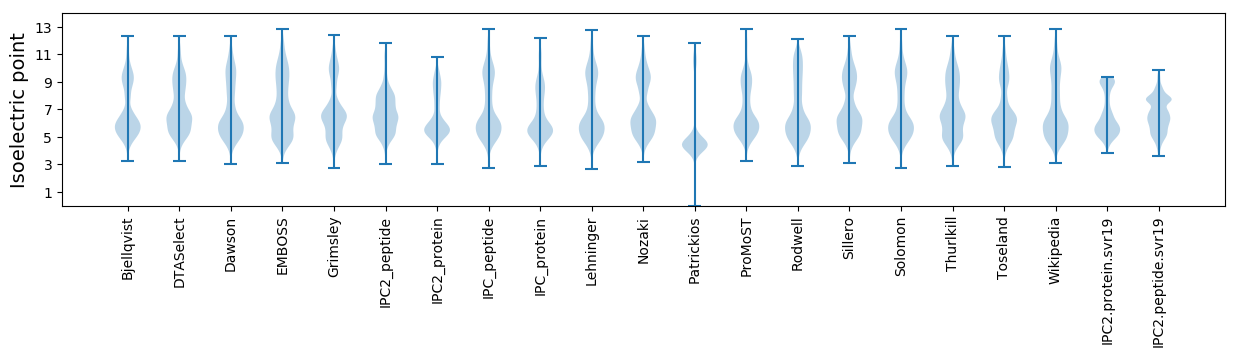
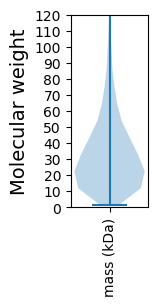
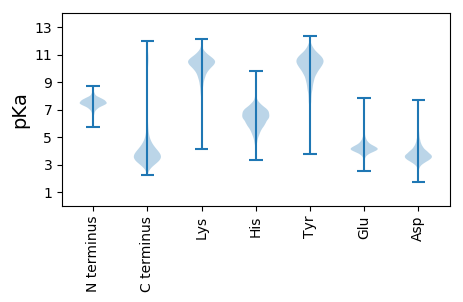
Protein with the lowest isoelectric point:
>tr|V6F5M1|V6F5M1_MAGGM High-affinity branched-chain amino acid transport protein (ABC superfamily ATP-binding) OS=Magnetospirillum gryphiswaldense (strain DSM 6361 / JCM 21280 / NBRC 15271 / MSR-1) OX=431944 GN=livG PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 10.13KK3 pKa = 9.86ILVASTALVAAGMITAGSAAASEE26 pKa = 4.44KK27 pKa = 10.55IKK29 pKa = 11.14LNLGGYY35 pKa = 7.39SKK37 pKa = 9.79WWVVGAWQDD46 pKa = 4.02DD47 pKa = 4.27SFTDD51 pKa = 5.48GINNTTAGQGDD62 pKa = 4.52ANSVDD67 pKa = 3.49VKK69 pKa = 11.29GEE71 pKa = 3.97NEE73 pKa = 3.84VWFGGSTTLDD83 pKa = 2.93NGLQVGIDD91 pKa = 3.38MQLRR95 pKa = 11.84AGGTTDD101 pKa = 3.33TTNTTGDD108 pKa = 4.19TIDD111 pKa = 4.25EE112 pKa = 4.58SYY114 pKa = 11.32VWISGGFGKK123 pKa = 10.85VIVGTEE129 pKa = 3.83NNGTYY134 pKa = 10.21LLHH137 pKa = 5.62VTAPDD142 pKa = 3.72AAGNWNEE149 pKa = 4.12GGVMMGNLAVARR161 pKa = 11.84PSAVSTLPGGNTTAILTDD179 pKa = 3.52GDD181 pKa = 4.19AEE183 pKa = 4.63KK184 pKa = 9.42ITYY187 pKa = 7.61VAPAFYY193 pKa = 10.75GLTLGATYY201 pKa = 10.62VPNATEE207 pKa = 4.65DD208 pKa = 3.67NQNVWGTGTPGTDD221 pKa = 2.88NQAAEE226 pKa = 4.27IYY228 pKa = 10.68GVGALYY234 pKa = 10.94ANTFGGVGVKK244 pKa = 9.9VSAGWVTYY252 pKa = 10.47DD253 pKa = 3.58IAQATARR260 pKa = 11.84SQEE263 pKa = 4.26VAFGTQLSYY272 pKa = 11.59AGFTLGGSYY281 pKa = 10.54RR282 pKa = 11.84DD283 pKa = 3.43VSQNANGGVNSVANANTASGYY304 pKa = 10.06GWDD307 pKa = 4.14VGLQYY312 pKa = 10.77ATGPYY317 pKa = 9.72AVSLAYY323 pKa = 10.41FNSKK327 pKa = 10.3VNGLTTTVDD336 pKa = 3.53DD337 pKa = 5.45DD338 pKa = 5.15EE339 pKa = 4.39ITAYY343 pKa = 10.35QLSGKK348 pKa = 9.53YY349 pKa = 10.63NMGAGVDD356 pKa = 3.72VLASIGHH363 pKa = 6.43IEE365 pKa = 4.38YY366 pKa = 10.44DD367 pKa = 3.74DD368 pKa = 4.41EE369 pKa = 4.68SAKK372 pKa = 8.12TTAADD377 pKa = 3.6ANHH380 pKa = 6.33NEE382 pKa = 4.09GWTVMTGLSLQFF394 pKa = 4.08
MM1 pKa = 7.45KK2 pKa = 10.13KK3 pKa = 9.86ILVASTALVAAGMITAGSAAASEE26 pKa = 4.44KK27 pKa = 10.55IKK29 pKa = 11.14LNLGGYY35 pKa = 7.39SKK37 pKa = 9.79WWVVGAWQDD46 pKa = 4.02DD47 pKa = 4.27SFTDD51 pKa = 5.48GINNTTAGQGDD62 pKa = 4.52ANSVDD67 pKa = 3.49VKK69 pKa = 11.29GEE71 pKa = 3.97NEE73 pKa = 3.84VWFGGSTTLDD83 pKa = 2.93NGLQVGIDD91 pKa = 3.38MQLRR95 pKa = 11.84AGGTTDD101 pKa = 3.33TTNTTGDD108 pKa = 4.19TIDD111 pKa = 4.25EE112 pKa = 4.58SYY114 pKa = 11.32VWISGGFGKK123 pKa = 10.85VIVGTEE129 pKa = 3.83NNGTYY134 pKa = 10.21LLHH137 pKa = 5.62VTAPDD142 pKa = 3.72AAGNWNEE149 pKa = 4.12GGVMMGNLAVARR161 pKa = 11.84PSAVSTLPGGNTTAILTDD179 pKa = 3.52GDD181 pKa = 4.19AEE183 pKa = 4.63KK184 pKa = 9.42ITYY187 pKa = 7.61VAPAFYY193 pKa = 10.75GLTLGATYY201 pKa = 10.62VPNATEE207 pKa = 4.65DD208 pKa = 3.67NQNVWGTGTPGTDD221 pKa = 2.88NQAAEE226 pKa = 4.27IYY228 pKa = 10.68GVGALYY234 pKa = 10.94ANTFGGVGVKK244 pKa = 9.9VSAGWVTYY252 pKa = 10.47DD253 pKa = 3.58IAQATARR260 pKa = 11.84SQEE263 pKa = 4.26VAFGTQLSYY272 pKa = 11.59AGFTLGGSYY281 pKa = 10.54RR282 pKa = 11.84DD283 pKa = 3.43VSQNANGGVNSVANANTASGYY304 pKa = 10.06GWDD307 pKa = 4.14VGLQYY312 pKa = 10.77ATGPYY317 pKa = 9.72AVSLAYY323 pKa = 10.41FNSKK327 pKa = 10.3VNGLTTTVDD336 pKa = 3.53DD337 pKa = 5.45DD338 pKa = 5.15EE339 pKa = 4.39ITAYY343 pKa = 10.35QLSGKK348 pKa = 9.53YY349 pKa = 10.63NMGAGVDD356 pKa = 3.72VLASIGHH363 pKa = 6.43IEE365 pKa = 4.38YY366 pKa = 10.44DD367 pKa = 3.74DD368 pKa = 4.41EE369 pKa = 4.68SAKK372 pKa = 8.12TTAADD377 pKa = 3.6ANHH380 pKa = 6.33NEE382 pKa = 4.09GWTVMTGLSLQFF394 pKa = 4.08
Molecular weight: 40.69 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|V6EWA5|V6EWA5_MAGGM Histidine kinase OS=Magnetospirillum gryphiswaldense (strain DSM 6361 / JCM 21280 / NBRC 15271 / MSR-1) OX=431944 GN=MGMSRv2__0350 PE=3 SV=1
MM1 pKa = 8.1DD2 pKa = 5.13IDD4 pKa = 3.46RR5 pKa = 11.84HH6 pKa = 5.39PRR8 pKa = 11.84NFHH11 pKa = 5.96GPSPGWKK18 pKa = 9.6RR19 pKa = 11.84LLCWRR24 pKa = 11.84HH25 pKa = 5.49LSFPGFPGFPGFPAPGLHH43 pKa = 6.0MGKK46 pKa = 9.99VSGPTQEE53 pKa = 4.16QAASLSLYY61 pKa = 10.69AEE63 pKa = 4.32TGGTRR68 pKa = 11.84DD69 pKa = 3.12RR70 pKa = 11.84HH71 pKa = 5.8EE72 pKa = 4.57DD73 pKa = 3.09QGLPVYY79 pKa = 10.35LAGEE83 pKa = 4.23RR84 pKa = 11.84AVEE87 pKa = 3.99RR88 pKa = 11.84CGMQLTAPTVKK99 pKa = 9.92RR100 pKa = 11.84GKK102 pKa = 9.76RR103 pKa = 11.84ALRR106 pKa = 11.84TPVRR110 pKa = 11.84AEE112 pKa = 3.29ITPRR116 pKa = 11.84SEE118 pKa = 3.45PRR120 pKa = 11.84PARR123 pKa = 11.84GAALPHH129 pKa = 6.71RR130 pKa = 11.84SGAEE134 pKa = 3.77PALSPHH140 pKa = 6.51LL141 pKa = 4.16
MM1 pKa = 8.1DD2 pKa = 5.13IDD4 pKa = 3.46RR5 pKa = 11.84HH6 pKa = 5.39PRR8 pKa = 11.84NFHH11 pKa = 5.96GPSPGWKK18 pKa = 9.6RR19 pKa = 11.84LLCWRR24 pKa = 11.84HH25 pKa = 5.49LSFPGFPGFPGFPAPGLHH43 pKa = 6.0MGKK46 pKa = 9.99VSGPTQEE53 pKa = 4.16QAASLSLYY61 pKa = 10.69AEE63 pKa = 4.32TGGTRR68 pKa = 11.84DD69 pKa = 3.12RR70 pKa = 11.84HH71 pKa = 5.8EE72 pKa = 4.57DD73 pKa = 3.09QGLPVYY79 pKa = 10.35LAGEE83 pKa = 4.23RR84 pKa = 11.84AVEE87 pKa = 3.99RR88 pKa = 11.84CGMQLTAPTVKK99 pKa = 9.92RR100 pKa = 11.84GKK102 pKa = 9.76RR103 pKa = 11.84ALRR106 pKa = 11.84TPVRR110 pKa = 11.84AEE112 pKa = 3.29ITPRR116 pKa = 11.84SEE118 pKa = 3.45PRR120 pKa = 11.84PARR123 pKa = 11.84GAALPHH129 pKa = 6.71RR130 pKa = 11.84SGAEE134 pKa = 3.77PALSPHH140 pKa = 6.51LL141 pKa = 4.16
Molecular weight: 15.4 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1329743 |
11 |
10342 |
320.0 |
34.68 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.56 ± 0.064 | 0.955 ± 0.018 |
6.043 ± 0.04 | 5.331 ± 0.041 |
3.416 ± 0.023 | 8.486 ± 0.044 |
2.24 ± 0.022 | 4.941 ± 0.025 |
3.468 ± 0.034 | 10.514 ± 0.061 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.651 ± 0.024 | 2.63 ± 0.038 |
5.014 ± 0.044 | 3.467 ± 0.024 |
6.841 ± 0.063 | 5.216 ± 0.037 |
5.244 ± 0.068 | 7.603 ± 0.032 |
1.366 ± 0.017 | 2.014 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
