
Streptomyces phage Coruscant
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; unclassified Siphoviridae
Average proteome isoelectric point is 6.33
Get precalculated fractions of proteins
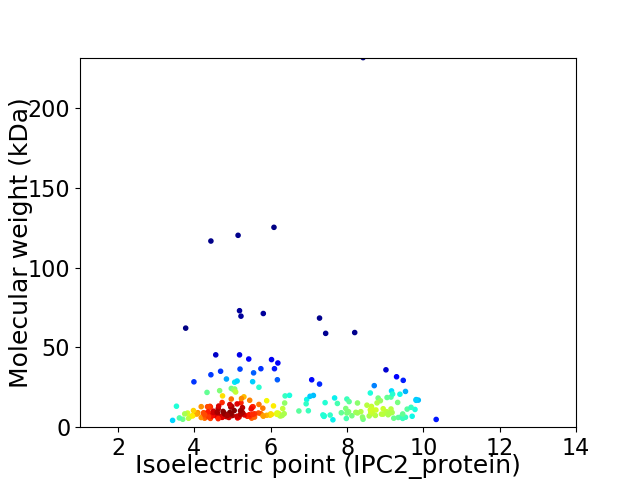
Virtual 2D-PAGE plot for 225 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A7G4AVY8|A0A7G4AVY8_9CAUD Uncharacterized protein OS=Streptomyces phage Coruscant OX=2739834 GN=HUN41_00049 PE=4 SV=1
MM1 pKa = 7.82SYY3 pKa = 10.56QLQVQADD10 pKa = 3.86APFSFWKK17 pKa = 10.48LDD19 pKa = 3.57GSGPIYY25 pKa = 10.36DD26 pKa = 4.9DD27 pKa = 3.31SAGSLRR33 pKa = 11.84QADD36 pKa = 4.07LVGTAVLSSALVAGSGNSLVLSNTNRR62 pKa = 11.84IEE64 pKa = 3.73MDD66 pKa = 3.31DD67 pKa = 3.53PVFNKK72 pKa = 10.37GYY74 pKa = 6.9EE75 pKa = 3.95QRR77 pKa = 11.84AFTLEE82 pKa = 3.89AWVKK86 pKa = 9.94PIDD89 pKa = 3.64VTGPVSVMSHH99 pKa = 5.6SSTYY103 pKa = 10.43DD104 pKa = 3.12GLVITEE110 pKa = 4.45DD111 pKa = 3.8EE112 pKa = 4.17ILFTTKK118 pKa = 10.48YY119 pKa = 10.26VGFGSCVASFRR130 pKa = 11.84YY131 pKa = 7.61TVNKK135 pKa = 9.31SYY137 pKa = 10.99HH138 pKa = 5.31VVGVHH143 pKa = 5.49NNGRR147 pKa = 11.84NSLYY151 pKa = 10.62VDD153 pKa = 3.88GVLVAEE159 pKa = 4.47TDD161 pKa = 3.54LTEE164 pKa = 4.5DD165 pKa = 3.38QQSAVYY171 pKa = 10.65DD172 pKa = 3.96FLTTDD177 pKa = 5.0LIAGQSGTASTISFDD192 pKa = 3.48APAIYY197 pKa = 10.51VGTLQSSIIEE207 pKa = 3.86SHH209 pKa = 6.07YY210 pKa = 10.95ASGVNVPSSQTIAYY224 pKa = 8.04YY225 pKa = 10.38NQGTFWNFSDD235 pKa = 3.72EE236 pKa = 3.9ARR238 pKa = 11.84NIGMTKK244 pKa = 8.19TWNLNAEE251 pKa = 4.24WQEE254 pKa = 3.93GTLTDD259 pKa = 3.59TVVANDD265 pKa = 4.48TIVPFYY271 pKa = 9.14TQSEE275 pKa = 4.79STVVVDD281 pKa = 4.55GLPTIEE287 pKa = 4.15FTNTSVAGTWQSSLEE302 pKa = 4.03LSGAPEE308 pKa = 3.77VTLAEE313 pKa = 5.04GRR315 pKa = 11.84LLWYY319 pKa = 10.8GEE321 pKa = 3.99GDD323 pKa = 3.51FTIEE327 pKa = 3.87YY328 pKa = 10.49SLDD331 pKa = 3.4GLVWNPPADD340 pKa = 3.7LATEE344 pKa = 4.3SLTNTDD350 pKa = 3.91SIDD353 pKa = 3.39VRR355 pKa = 11.84VSFTGGIVDD364 pKa = 4.6DD365 pKa = 4.85PSFVDD370 pKa = 3.81YY371 pKa = 11.12INLVVYY377 pKa = 9.22TDD379 pKa = 3.48KK380 pKa = 11.57DD381 pKa = 4.26FIGSLDD387 pKa = 3.39NRR389 pKa = 11.84FASFTGTGVTSQNYY403 pKa = 9.08FEE405 pKa = 5.43PIEE408 pKa = 4.0YY409 pKa = 10.73ADD411 pKa = 3.79FNGIKK416 pKa = 9.8IATGEE421 pKa = 3.99ITITTDD427 pKa = 2.54ASFNGEE433 pKa = 4.43EE434 pKa = 4.17EE435 pKa = 4.48TPDD438 pKa = 3.26QLFITGFDD446 pKa = 3.47LWAKK450 pKa = 9.9AAVGNIVVAGANTITRR466 pKa = 11.84SGDD469 pKa = 3.48TIVFSGFDD477 pKa = 3.1AVTVNGEE484 pKa = 4.21LVTTGATVFGNDD496 pKa = 2.9DD497 pKa = 3.39WMHH500 pKa = 6.39IGAVLTTPLNPTITVGDD517 pKa = 4.2DD518 pKa = 3.44GASVAQFSALYY529 pKa = 10.72GEE531 pKa = 4.39VDD533 pKa = 3.54GTGMQQIYY541 pKa = 10.13AAYY544 pKa = 9.32LGLPGVVVEE553 pKa = 5.24DD554 pKa = 4.02SSTVTISQPANPTKK568 pKa = 10.39LYY570 pKa = 10.59SNIWGITPAGG580 pKa = 3.46
MM1 pKa = 7.82SYY3 pKa = 10.56QLQVQADD10 pKa = 3.86APFSFWKK17 pKa = 10.48LDD19 pKa = 3.57GSGPIYY25 pKa = 10.36DD26 pKa = 4.9DD27 pKa = 3.31SAGSLRR33 pKa = 11.84QADD36 pKa = 4.07LVGTAVLSSALVAGSGNSLVLSNTNRR62 pKa = 11.84IEE64 pKa = 3.73MDD66 pKa = 3.31DD67 pKa = 3.53PVFNKK72 pKa = 10.37GYY74 pKa = 6.9EE75 pKa = 3.95QRR77 pKa = 11.84AFTLEE82 pKa = 3.89AWVKK86 pKa = 9.94PIDD89 pKa = 3.64VTGPVSVMSHH99 pKa = 5.6SSTYY103 pKa = 10.43DD104 pKa = 3.12GLVITEE110 pKa = 4.45DD111 pKa = 3.8EE112 pKa = 4.17ILFTTKK118 pKa = 10.48YY119 pKa = 10.26VGFGSCVASFRR130 pKa = 11.84YY131 pKa = 7.61TVNKK135 pKa = 9.31SYY137 pKa = 10.99HH138 pKa = 5.31VVGVHH143 pKa = 5.49NNGRR147 pKa = 11.84NSLYY151 pKa = 10.62VDD153 pKa = 3.88GVLVAEE159 pKa = 4.47TDD161 pKa = 3.54LTEE164 pKa = 4.5DD165 pKa = 3.38QQSAVYY171 pKa = 10.65DD172 pKa = 3.96FLTTDD177 pKa = 5.0LIAGQSGTASTISFDD192 pKa = 3.48APAIYY197 pKa = 10.51VGTLQSSIIEE207 pKa = 3.86SHH209 pKa = 6.07YY210 pKa = 10.95ASGVNVPSSQTIAYY224 pKa = 8.04YY225 pKa = 10.38NQGTFWNFSDD235 pKa = 3.72EE236 pKa = 3.9ARR238 pKa = 11.84NIGMTKK244 pKa = 8.19TWNLNAEE251 pKa = 4.24WQEE254 pKa = 3.93GTLTDD259 pKa = 3.59TVVANDD265 pKa = 4.48TIVPFYY271 pKa = 9.14TQSEE275 pKa = 4.79STVVVDD281 pKa = 4.55GLPTIEE287 pKa = 4.15FTNTSVAGTWQSSLEE302 pKa = 4.03LSGAPEE308 pKa = 3.77VTLAEE313 pKa = 5.04GRR315 pKa = 11.84LLWYY319 pKa = 10.8GEE321 pKa = 3.99GDD323 pKa = 3.51FTIEE327 pKa = 3.87YY328 pKa = 10.49SLDD331 pKa = 3.4GLVWNPPADD340 pKa = 3.7LATEE344 pKa = 4.3SLTNTDD350 pKa = 3.91SIDD353 pKa = 3.39VRR355 pKa = 11.84VSFTGGIVDD364 pKa = 4.6DD365 pKa = 4.85PSFVDD370 pKa = 3.81YY371 pKa = 11.12INLVVYY377 pKa = 9.22TDD379 pKa = 3.48KK380 pKa = 11.57DD381 pKa = 4.26FIGSLDD387 pKa = 3.39NRR389 pKa = 11.84FASFTGTGVTSQNYY403 pKa = 9.08FEE405 pKa = 5.43PIEE408 pKa = 4.0YY409 pKa = 10.73ADD411 pKa = 3.79FNGIKK416 pKa = 9.8IATGEE421 pKa = 3.99ITITTDD427 pKa = 2.54ASFNGEE433 pKa = 4.43EE434 pKa = 4.17EE435 pKa = 4.48TPDD438 pKa = 3.26QLFITGFDD446 pKa = 3.47LWAKK450 pKa = 9.9AAVGNIVVAGANTITRR466 pKa = 11.84SGDD469 pKa = 3.48TIVFSGFDD477 pKa = 3.1AVTVNGEE484 pKa = 4.21LVTTGATVFGNDD496 pKa = 2.9DD497 pKa = 3.39WMHH500 pKa = 6.39IGAVLTTPLNPTITVGDD517 pKa = 4.2DD518 pKa = 3.44GASVAQFSALYY529 pKa = 10.72GEE531 pKa = 4.39VDD533 pKa = 3.54GTGMQQIYY541 pKa = 10.13AAYY544 pKa = 9.32LGLPGVVVEE553 pKa = 5.24DD554 pKa = 4.02SSTVTISQPANPTKK568 pKa = 10.39LYY570 pKa = 10.59SNIWGITPAGG580 pKa = 3.46
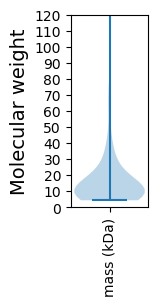
Molecular weight: 62.11 kDa
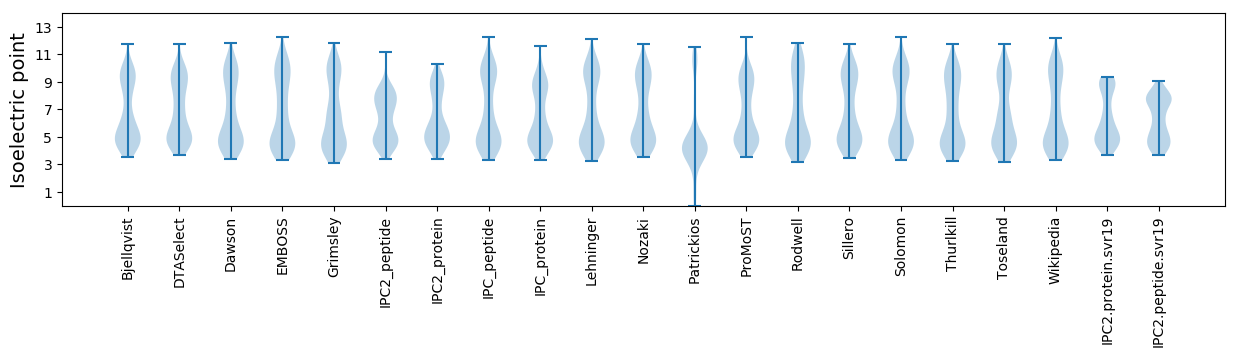
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A7G4AVY0|A0A7G4AVY0_9CAUD Uncharacterized protein OS=Streptomyces phage Coruscant OX=2739834 GN=HUN41_00041 PE=4 SV=1
MM1 pKa = 7.58KK2 pKa = 10.26SDD4 pKa = 3.61EE5 pKa = 4.28DD6 pKa = 3.36RR7 pKa = 11.84CRR9 pKa = 11.84EE10 pKa = 3.86YY11 pKa = 11.06GVPYY15 pKa = 10.08DD16 pKa = 3.83PQITRR21 pKa = 11.84QEE23 pKa = 4.35VYY25 pKa = 10.5DD26 pKa = 3.75KK27 pKa = 11.38SRR29 pKa = 11.84WKK31 pKa = 10.74CHH33 pKa = 4.55ICGKK37 pKa = 9.32RR38 pKa = 11.84VYY40 pKa = 10.06RR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84KK44 pKa = 8.32YY45 pKa = 9.51PHH47 pKa = 7.17PKK49 pKa = 9.74SASLDD54 pKa = 3.67HH55 pKa = 6.25VVPLSWRR62 pKa = 11.84KK63 pKa = 9.74KK64 pKa = 9.55SPGHH68 pKa = 4.51VWGNVALAHH77 pKa = 6.11LVCNQRR83 pKa = 11.84KK84 pKa = 7.68GARR87 pKa = 11.84FAGSKK92 pKa = 10.02RR93 pKa = 11.84RR94 pKa = 11.84IPKK97 pKa = 9.51KK98 pKa = 10.43YY99 pKa = 10.49SISTVNLFRR108 pKa = 11.84LTIFSITALAFYY120 pKa = 9.07WEE122 pKa = 5.03CPPEE126 pKa = 4.0ILITGAGLCILSVTRR141 pKa = 11.84RR142 pKa = 11.84PRR144 pKa = 11.84RR145 pKa = 11.84RR146 pKa = 11.84RR147 pKa = 11.84AA148 pKa = 2.96
MM1 pKa = 7.58KK2 pKa = 10.26SDD4 pKa = 3.61EE5 pKa = 4.28DD6 pKa = 3.36RR7 pKa = 11.84CRR9 pKa = 11.84EE10 pKa = 3.86YY11 pKa = 11.06GVPYY15 pKa = 10.08DD16 pKa = 3.83PQITRR21 pKa = 11.84QEE23 pKa = 4.35VYY25 pKa = 10.5DD26 pKa = 3.75KK27 pKa = 11.38SRR29 pKa = 11.84WKK31 pKa = 10.74CHH33 pKa = 4.55ICGKK37 pKa = 9.32RR38 pKa = 11.84VYY40 pKa = 10.06RR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84KK44 pKa = 8.32YY45 pKa = 9.51PHH47 pKa = 7.17PKK49 pKa = 9.74SASLDD54 pKa = 3.67HH55 pKa = 6.25VVPLSWRR62 pKa = 11.84KK63 pKa = 9.74KK64 pKa = 9.55SPGHH68 pKa = 4.51VWGNVALAHH77 pKa = 6.11LVCNQRR83 pKa = 11.84KK84 pKa = 7.68GARR87 pKa = 11.84FAGSKK92 pKa = 10.02RR93 pKa = 11.84RR94 pKa = 11.84IPKK97 pKa = 9.51KK98 pKa = 10.43YY99 pKa = 10.49SISTVNLFRR108 pKa = 11.84LTIFSITALAFYY120 pKa = 9.07WEE122 pKa = 5.03CPPEE126 pKa = 4.0ILITGAGLCILSVTRR141 pKa = 11.84RR142 pKa = 11.84PRR144 pKa = 11.84RR145 pKa = 11.84RR146 pKa = 11.84RR147 pKa = 11.84AA148 pKa = 2.96
Molecular weight: 17.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
34808 |
38 |
2133 |
154.7 |
17.42 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.969 ± 0.37 | 1.135 ± 0.114 |
6.323 ± 0.169 | 6.996 ± 0.242 |
4.24 ± 0.109 | 7.228 ± 0.169 |
1.913 ± 0.125 | 5.685 ± 0.131 |
6.343 ± 0.244 | 7.021 ± 0.152 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.982 ± 0.135 | 4.72 ± 0.168 |
3.66 ± 0.164 | 3.353 ± 0.202 |
5.576 ± 0.16 | 6.162 ± 0.212 |
6.036 ± 0.214 | 7.079 ± 0.192 |
1.856 ± 0.088 | 3.72 ± 0.165 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
