
Methanosarcinales archaeon
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Methanomicrobia; Methanosarcinales; unclassified Methanosarcinales
Average proteome isoelectric point is 7.0
Get precalculated fractions of proteins
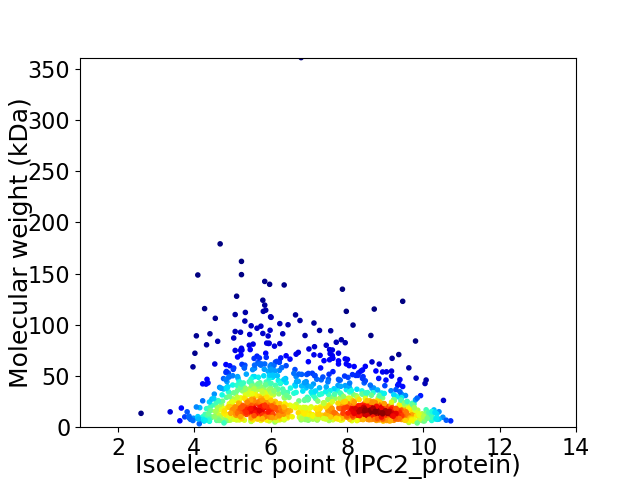
Virtual 2D-PAGE plot for 1193 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A482R572|A0A482R572_9EURY Uncharacterized protein OS=Methanosarcinales archaeon OX=2250255 GN=EOO65_03780 PE=4 SV=1
MM1 pKa = 7.35LSSYY5 pKa = 11.08ACWSLSYY12 pKa = 9.73TVLSITPLAAKK23 pKa = 9.97CQTPTNLFTYY33 pKa = 10.4SKK35 pKa = 9.3TDD37 pKa = 2.76NRR39 pKa = 11.84LRR41 pKa = 11.84IGATPLRR48 pKa = 11.84AYY50 pKa = 10.58NCTRR54 pKa = 11.84GYY56 pKa = 9.3TVVLRR61 pKa = 11.84AYY63 pKa = 7.37RR64 pKa = 11.84TSYY67 pKa = 7.99PTNFTDD73 pKa = 5.24CRR75 pKa = 11.84IEE77 pKa = 4.58LAVQQTNEE85 pKa = 4.37PPSFLPGACAPRR97 pKa = 11.84SVPEE101 pKa = 3.88RR102 pKa = 11.84ADD104 pKa = 2.83IGTVVGGVVAATDD117 pKa = 4.07PNVGQHH123 pKa = 6.03LVWSIDD129 pKa = 3.27AGAVIYY135 pKa = 9.91PFDD138 pKa = 4.55IGPCTGIITVNDD150 pKa = 3.71GAINYY155 pKa = 7.35TRR157 pKa = 11.84NAVFTLPVRR166 pKa = 11.84VMDD169 pKa = 3.89VGLTPSMSALCSITINVIDD188 pKa = 4.51VNDD191 pKa = 4.38PPTILTTEE199 pKa = 4.3LSAFEE204 pKa = 4.14NSDD207 pKa = 3.19VGEE210 pKa = 4.14EE211 pKa = 3.93VGQIVVFDD219 pKa = 5.01PDD221 pKa = 3.45TAGANLTYY229 pKa = 10.85NWTLVDD235 pKa = 3.81SSDD238 pKa = 3.12AFAISRR244 pKa = 11.84SGMVTVKK251 pKa = 10.53RR252 pKa = 11.84DD253 pKa = 3.31ILDD256 pKa = 3.94FEE258 pKa = 4.74VKK260 pKa = 8.45PTYY263 pKa = 9.26TYY265 pKa = 10.84RR266 pKa = 11.84VGVFDD271 pKa = 4.38GDD273 pKa = 3.6NFEE276 pKa = 4.65EE277 pKa = 4.87GVVVITLMDD286 pKa = 4.69ANDD289 pKa = 4.52PPTVHH294 pKa = 7.08PGSFSVPEE302 pKa = 3.94MSPAGTLVSGGSITVTDD319 pKa = 3.59QDD321 pKa = 3.19GDD323 pKa = 3.7AVTFEE328 pKa = 4.41VPVVDD333 pKa = 3.99TFTISGGRR341 pKa = 11.84FYY343 pKa = 11.34VAAGVVLDD351 pKa = 4.24YY352 pKa = 8.0EE353 pKa = 4.66TTPSYY358 pKa = 11.17SVDD361 pKa = 3.72VIANDD366 pKa = 3.54ANEE369 pKa = 4.13ASTVASITIRR379 pKa = 11.84VIDD382 pKa = 3.87VNEE385 pKa = 3.89VPEE388 pKa = 4.17FDD390 pKa = 3.84EE391 pKa = 4.5ATGVAMSIEE400 pKa = 4.71EE401 pKa = 4.08DD402 pKa = 3.96TLPGDD407 pKa = 3.87ALGAPISASDD417 pKa = 3.84PDD419 pKa = 4.16ANTILTYY426 pKa = 10.48EE427 pKa = 4.25IVTVTPAGNNHH438 pKa = 5.45QFSIDD443 pKa = 3.4GLTGQLRR450 pKa = 11.84AKK452 pKa = 8.4STSYY456 pKa = 10.86FDD458 pKa = 3.78YY459 pKa = 10.59EE460 pKa = 4.48LGVRR464 pKa = 11.84TYY466 pKa = 9.44TLAVKK471 pKa = 9.17VTDD474 pKa = 3.58NGSPRR479 pKa = 11.84LSATTTVTVTVTNVNEE495 pKa = 4.11APAFSAASYY504 pKa = 7.67TWSVRR509 pKa = 11.84EE510 pKa = 3.97DD511 pKa = 3.81TVVGTVMGSVFATDD525 pKa = 3.76PEE527 pKa = 4.86GTAVTYY533 pKa = 10.48EE534 pKa = 4.09LTTPSSLVTMSASGAFSVATDD555 pKa = 3.54LL556 pKa = 5.15
MM1 pKa = 7.35LSSYY5 pKa = 11.08ACWSLSYY12 pKa = 9.73TVLSITPLAAKK23 pKa = 9.97CQTPTNLFTYY33 pKa = 10.4SKK35 pKa = 9.3TDD37 pKa = 2.76NRR39 pKa = 11.84LRR41 pKa = 11.84IGATPLRR48 pKa = 11.84AYY50 pKa = 10.58NCTRR54 pKa = 11.84GYY56 pKa = 9.3TVVLRR61 pKa = 11.84AYY63 pKa = 7.37RR64 pKa = 11.84TSYY67 pKa = 7.99PTNFTDD73 pKa = 5.24CRR75 pKa = 11.84IEE77 pKa = 4.58LAVQQTNEE85 pKa = 4.37PPSFLPGACAPRR97 pKa = 11.84SVPEE101 pKa = 3.88RR102 pKa = 11.84ADD104 pKa = 2.83IGTVVGGVVAATDD117 pKa = 4.07PNVGQHH123 pKa = 6.03LVWSIDD129 pKa = 3.27AGAVIYY135 pKa = 9.91PFDD138 pKa = 4.55IGPCTGIITVNDD150 pKa = 3.71GAINYY155 pKa = 7.35TRR157 pKa = 11.84NAVFTLPVRR166 pKa = 11.84VMDD169 pKa = 3.89VGLTPSMSALCSITINVIDD188 pKa = 4.51VNDD191 pKa = 4.38PPTILTTEE199 pKa = 4.3LSAFEE204 pKa = 4.14NSDD207 pKa = 3.19VGEE210 pKa = 4.14EE211 pKa = 3.93VGQIVVFDD219 pKa = 5.01PDD221 pKa = 3.45TAGANLTYY229 pKa = 10.85NWTLVDD235 pKa = 3.81SSDD238 pKa = 3.12AFAISRR244 pKa = 11.84SGMVTVKK251 pKa = 10.53RR252 pKa = 11.84DD253 pKa = 3.31ILDD256 pKa = 3.94FEE258 pKa = 4.74VKK260 pKa = 8.45PTYY263 pKa = 9.26TYY265 pKa = 10.84RR266 pKa = 11.84VGVFDD271 pKa = 4.38GDD273 pKa = 3.6NFEE276 pKa = 4.65EE277 pKa = 4.87GVVVITLMDD286 pKa = 4.69ANDD289 pKa = 4.52PPTVHH294 pKa = 7.08PGSFSVPEE302 pKa = 3.94MSPAGTLVSGGSITVTDD319 pKa = 3.59QDD321 pKa = 3.19GDD323 pKa = 3.7AVTFEE328 pKa = 4.41VPVVDD333 pKa = 3.99TFTISGGRR341 pKa = 11.84FYY343 pKa = 11.34VAAGVVLDD351 pKa = 4.24YY352 pKa = 8.0EE353 pKa = 4.66TTPSYY358 pKa = 11.17SVDD361 pKa = 3.72VIANDD366 pKa = 3.54ANEE369 pKa = 4.13ASTVASITIRR379 pKa = 11.84VIDD382 pKa = 3.87VNEE385 pKa = 3.89VPEE388 pKa = 4.17FDD390 pKa = 3.84EE391 pKa = 4.5ATGVAMSIEE400 pKa = 4.71EE401 pKa = 4.08DD402 pKa = 3.96TLPGDD407 pKa = 3.87ALGAPISASDD417 pKa = 3.84PDD419 pKa = 4.16ANTILTYY426 pKa = 10.48EE427 pKa = 4.25IVTVTPAGNNHH438 pKa = 5.45QFSIDD443 pKa = 3.4GLTGQLRR450 pKa = 11.84AKK452 pKa = 8.4STSYY456 pKa = 10.86FDD458 pKa = 3.78YY459 pKa = 10.59EE460 pKa = 4.48LGVRR464 pKa = 11.84TYY466 pKa = 9.44TLAVKK471 pKa = 9.17VTDD474 pKa = 3.58NGSPRR479 pKa = 11.84LSATTTVTVTVTNVNEE495 pKa = 4.11APAFSAASYY504 pKa = 7.67TWSVRR509 pKa = 11.84EE510 pKa = 3.97DD511 pKa = 3.81TVVGTVMGSVFATDD525 pKa = 3.76PEE527 pKa = 4.86GTAVTYY533 pKa = 10.48EE534 pKa = 4.09LTTPSSLVTMSASGAFSVATDD555 pKa = 3.54LL556 pKa = 5.15
Molecular weight: 58.92 kDa
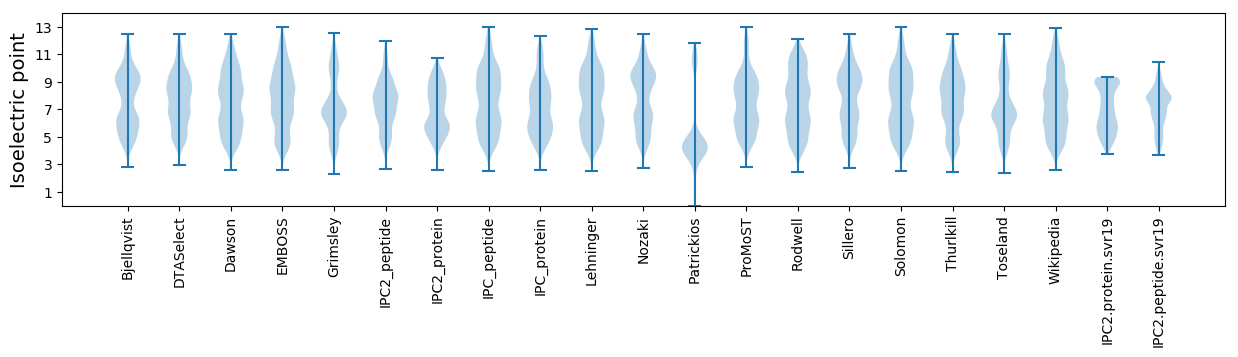
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A482R4R5|A0A482R4R5_9EURY Uncharacterized protein OS=Methanosarcinales archaeon OX=2250255 GN=EOO65_00195 PE=4 SV=1
MM1 pKa = 6.96VASATTSSAAAALARR16 pKa = 11.84WLRR19 pKa = 11.84TPRR22 pKa = 11.84SHH24 pKa = 7.39TPFLWGVHH32 pKa = 4.87VCVCLCARR40 pKa = 11.84VCIAWCAGAVVSASWDD56 pKa = 3.39ATVKK60 pKa = 10.28VWDD63 pKa = 4.13LRR65 pKa = 11.84SPSMAAPEE73 pKa = 4.35DD74 pKa = 3.76ASTRR78 pKa = 11.84AADD81 pKa = 3.47MTLALPEE88 pKa = 4.41RR89 pKa = 11.84AFSMSLCSDD98 pKa = 4.16YY99 pKa = 11.13IVLVATAGQHH109 pKa = 5.52LVAYY113 pKa = 8.89DD114 pKa = 3.7LRR116 pKa = 11.84KK117 pKa = 9.89GDD119 pKa = 3.33EE120 pKa = 4.26MLWKK124 pKa = 10.4RR125 pKa = 11.84EE126 pKa = 4.12STVKK130 pKa = 10.55FQLRR134 pKa = 11.84CIRR137 pKa = 11.84AFADD141 pKa = 3.46GKK143 pKa = 10.73GYY145 pKa = 10.51VVSSIDD151 pKa = 3.26GRR153 pKa = 11.84CGVEE157 pKa = 3.93YY158 pKa = 10.36MEE160 pKa = 4.91EE161 pKa = 4.11ADD163 pKa = 4.06GKK165 pKa = 9.55PFSFKK170 pKa = 9.97CHH172 pKa = 6.36RR173 pKa = 11.84SKK175 pKa = 10.05TPDD178 pKa = 3.1GKK180 pKa = 10.96EE181 pKa = 3.73LITPVNALAFNPVYY195 pKa = 10.14NTFVTGGGDD204 pKa = 3.48GTVAAWDD211 pKa = 3.79HH212 pKa = 5.58VSKK215 pKa = 10.89KK216 pKa = 10.19RR217 pKa = 11.84LAVLGTYY224 pKa = 7.32PTSIAALAFNRR235 pKa = 11.84CVQAGYY241 pKa = 10.82ASPARR246 pKa = 11.84PSARR250 pKa = 11.84ANAVVFTLPPRR261 pKa = 11.84VRR263 pKa = 11.84THH265 pKa = 5.23VLARR269 pKa = 11.84ACACVCVCVADD280 pKa = 4.42AGRR283 pKa = 11.84APSWRR288 pKa = 11.84LPRR291 pKa = 11.84RR292 pKa = 11.84TRR294 pKa = 11.84GRR296 pKa = 11.84RR297 pKa = 11.84VRR299 pKa = 11.84RR300 pKa = 11.84SIPRR304 pKa = 11.84TASMCTVWGAPWLARR319 pKa = 11.84ALRR322 pKa = 11.84KK323 pKa = 9.87VADD326 pKa = 3.8PAAMVVRR333 pKa = 11.84HH334 pKa = 5.67ASWRR338 pKa = 11.84RR339 pKa = 11.84CRR341 pKa = 11.84PSWRR345 pKa = 11.84ARR347 pKa = 11.84PRR349 pKa = 11.84VRR351 pKa = 11.84VVPLSLILRR360 pKa = 11.84TSLL363 pKa = 3.81
MM1 pKa = 6.96VASATTSSAAAALARR16 pKa = 11.84WLRR19 pKa = 11.84TPRR22 pKa = 11.84SHH24 pKa = 7.39TPFLWGVHH32 pKa = 4.87VCVCLCARR40 pKa = 11.84VCIAWCAGAVVSASWDD56 pKa = 3.39ATVKK60 pKa = 10.28VWDD63 pKa = 4.13LRR65 pKa = 11.84SPSMAAPEE73 pKa = 4.35DD74 pKa = 3.76ASTRR78 pKa = 11.84AADD81 pKa = 3.47MTLALPEE88 pKa = 4.41RR89 pKa = 11.84AFSMSLCSDD98 pKa = 4.16YY99 pKa = 11.13IVLVATAGQHH109 pKa = 5.52LVAYY113 pKa = 8.89DD114 pKa = 3.7LRR116 pKa = 11.84KK117 pKa = 9.89GDD119 pKa = 3.33EE120 pKa = 4.26MLWKK124 pKa = 10.4RR125 pKa = 11.84EE126 pKa = 4.12STVKK130 pKa = 10.55FQLRR134 pKa = 11.84CIRR137 pKa = 11.84AFADD141 pKa = 3.46GKK143 pKa = 10.73GYY145 pKa = 10.51VVSSIDD151 pKa = 3.26GRR153 pKa = 11.84CGVEE157 pKa = 3.93YY158 pKa = 10.36MEE160 pKa = 4.91EE161 pKa = 4.11ADD163 pKa = 4.06GKK165 pKa = 9.55PFSFKK170 pKa = 9.97CHH172 pKa = 6.36RR173 pKa = 11.84SKK175 pKa = 10.05TPDD178 pKa = 3.1GKK180 pKa = 10.96EE181 pKa = 3.73LITPVNALAFNPVYY195 pKa = 10.14NTFVTGGGDD204 pKa = 3.48GTVAAWDD211 pKa = 3.79HH212 pKa = 5.58VSKK215 pKa = 10.89KK216 pKa = 10.19RR217 pKa = 11.84LAVLGTYY224 pKa = 7.32PTSIAALAFNRR235 pKa = 11.84CVQAGYY241 pKa = 10.82ASPARR246 pKa = 11.84PSARR250 pKa = 11.84ANAVVFTLPPRR261 pKa = 11.84VRR263 pKa = 11.84THH265 pKa = 5.23VLARR269 pKa = 11.84ACACVCVCVADD280 pKa = 4.42AGRR283 pKa = 11.84APSWRR288 pKa = 11.84LPRR291 pKa = 11.84RR292 pKa = 11.84TRR294 pKa = 11.84GRR296 pKa = 11.84RR297 pKa = 11.84VRR299 pKa = 11.84RR300 pKa = 11.84SIPRR304 pKa = 11.84TASMCTVWGAPWLARR319 pKa = 11.84ALRR322 pKa = 11.84KK323 pKa = 9.87VADD326 pKa = 3.8PAAMVVRR333 pKa = 11.84HH334 pKa = 5.67ASWRR338 pKa = 11.84RR339 pKa = 11.84CRR341 pKa = 11.84PSWRR345 pKa = 11.84ARR347 pKa = 11.84PRR349 pKa = 11.84VRR351 pKa = 11.84VVPLSLILRR360 pKa = 11.84TSLL363 pKa = 3.81
Molecular weight: 39.7 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
315384 |
34 |
3471 |
264.4 |
28.39 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.02 ± 0.111 | 2.205 ± 0.051 |
4.771 ± 0.06 | 4.293 ± 0.083 |
3.099 ± 0.046 | 6.848 ± 0.109 |
2.824 ± 0.055 | 3.434 ± 0.05 |
2.571 ± 0.066 | 9.177 ± 0.089 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.5 ± 0.037 | 2.917 ± 0.052 |
5.626 ± 0.077 | 3.377 ± 0.047 |
6.582 ± 0.07 | 8.565 ± 0.113 |
6.651 ± 0.084 | 8.065 ± 0.075 |
1.356 ± 0.034 | 2.117 ± 0.045 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
