
Pseudomonas salegens
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Pseudomonadales; Pseudomonadaceae; Pseudomonas
Average proteome isoelectric point is 6.04
Get precalculated fractions of proteins
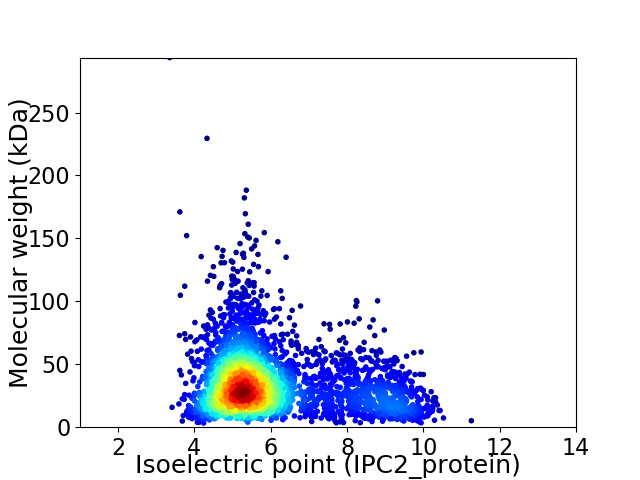
Virtual 2D-PAGE plot for 3414 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H2F372|A0A1H2F372_9PSED Thiol peroxidase OS=Pseudomonas salegens OX=1434072 GN=tpx PE=3 SV=1
MM1 pKa = 7.21TAGFSGTRR9 pKa = 11.84EE10 pKa = 3.69RR11 pKa = 11.84GFSFFRR17 pKa = 11.84GLAQSVPGKK26 pKa = 10.57KK27 pKa = 10.26LLTSLLAAPMLMLVAGFASAQSVDD51 pKa = 3.51LVLNHH56 pKa = 6.41GQDD59 pKa = 3.61LPTINATDD67 pKa = 3.54TVTFTVTVDD76 pKa = 3.14NSDD79 pKa = 3.51PGFTADD85 pKa = 3.74AANVVLDD92 pKa = 3.89YY93 pKa = 10.97QISPTDD99 pKa = 3.46TLVSVTPSTGCSAPDD114 pKa = 3.25ASNAFSCNLGTLLHH128 pKa = 6.18GTDD131 pKa = 3.32TSLDD135 pKa = 3.2IVVRR139 pKa = 11.84GTGTGVSNFPYY150 pKa = 10.76VMQTTATVSTSNNDD164 pKa = 2.78IDD166 pKa = 4.19PSNNTEE172 pKa = 3.87QRR174 pKa = 11.84NVTVNAGTDD183 pKa = 3.29LGVVLNSSPGMTPSGGAWTHH203 pKa = 6.97DD204 pKa = 2.81IVVDD208 pKa = 3.6NYY210 pKa = 11.46GPIDD214 pKa = 3.54ATNAVIHH221 pKa = 6.92LDD223 pKa = 3.67LPVGVVVQSVPAGCSVAAPGVVCQVGALSVGDD255 pKa = 4.31SLPLGSIVTQVAVASGSNLSATALIDD281 pKa = 5.02SLDD284 pKa = 3.67QYY286 pKa = 11.47DD287 pKa = 4.14GNSINQQSTRR297 pKa = 11.84QVEE300 pKa = 4.39VTPGSDD306 pKa = 2.95LSVGLSHH313 pKa = 7.1TAGTILEE320 pKa = 4.3GQPFNLTASPAYY332 pKa = 9.0TGEE335 pKa = 4.21VPDD338 pKa = 4.66IPALVIDD345 pKa = 4.83VDD347 pKa = 4.05PGLQILSPANFSQNGWSCSVTGQQVSCSGLDD378 pKa = 3.35VGSMVPGSSQSLGDD392 pKa = 3.32VSVQVSGATAGSFAANQAQISADD415 pKa = 3.52YY416 pKa = 10.53SVKK419 pKa = 10.21PDD421 pKa = 3.33SDD423 pKa = 3.53LSNNVAGTTPVIEE436 pKa = 4.28SANLNVGINKK446 pKa = 9.64SPPNRR451 pKa = 11.84NLRR454 pKa = 11.84TIGAAYY460 pKa = 9.25PFDD463 pKa = 3.56YY464 pKa = 10.59QIRR467 pKa = 11.84VRR469 pKa = 11.84NNGNIAFWGDD479 pKa = 3.73LTWSDD484 pKa = 4.03NVPAGLTVTAINGPAGWSCSPALPVTGAATIDD516 pKa = 3.68CVRR519 pKa = 11.84TYY521 pKa = 11.12TEE523 pKa = 4.24GAPLAVNATQDD534 pKa = 3.68FTVTAYY540 pKa = 10.74ANGAISGNITNQACLTQIDD559 pKa = 4.38HH560 pKa = 7.74DD561 pKa = 4.8LAFLDD566 pKa = 4.13NVDD569 pKa = 4.19PAPCNGATINAQALNDD585 pKa = 3.98SADD588 pKa = 3.53LRR590 pKa = 11.84VVKK593 pKa = 9.0TASAATVNAGEE604 pKa = 4.2ALTYY608 pKa = 8.13TLRR611 pKa = 11.84IVNDD615 pKa = 4.59GPATASDD622 pKa = 3.72VDD624 pKa = 3.98LTDD627 pKa = 3.7AFSALFTGSAGNSFQGATVTNATASGGTCGTSGLAGNVGSRR668 pKa = 11.84TLSCNFASIPVCSGTDD684 pKa = 3.39CPSIEE689 pKa = 4.41VTVLPLGNTSSGVPLVRR706 pKa = 11.84NNTASAYY713 pKa = 10.39SNEE716 pKa = 4.47TADD719 pKa = 4.09PDD721 pKa = 3.89YY722 pKa = 11.21GNNPGSVSTTINPLTDD738 pKa = 3.87LLVSKK743 pKa = 10.68DD744 pKa = 3.42VTVWSGNLGTPLEE757 pKa = 4.11YY758 pKa = 10.24RR759 pKa = 11.84LQIEE763 pKa = 4.71NIGASGASGLSLEE776 pKa = 5.2DD777 pKa = 3.84VLPHH781 pKa = 7.03DD782 pKa = 4.6LTYY785 pKa = 11.31LSTDD789 pKa = 3.29PGASASCATEE799 pKa = 3.93PGANTTTTTGNDD811 pKa = 3.26EE812 pKa = 4.23IACTRR817 pKa = 11.84STLPRR822 pKa = 11.84NATWTVTVNTRR833 pKa = 11.84PNHH836 pKa = 6.47GIPAGTTLVNSVDD849 pKa = 3.96AEE851 pKa = 4.3TDD853 pKa = 3.38TPEE856 pKa = 4.11TDD858 pKa = 3.54LSNNADD864 pKa = 3.6TADD867 pKa = 3.83AEE869 pKa = 4.33VGEE872 pKa = 4.51AVVDD876 pKa = 4.01LAVIKK881 pKa = 10.65ADD883 pKa = 3.53TPDD886 pKa = 3.5PVFVGDD892 pKa = 3.43RR893 pKa = 11.84VTYY896 pKa = 8.78SLRR899 pKa = 11.84IINNGPSVATDD910 pKa = 3.18ATIYY914 pKa = 10.67DD915 pKa = 4.32YY916 pKa = 11.37LPSAGLRR923 pKa = 11.84YY924 pKa = 9.89VEE926 pKa = 4.0NSIRR930 pKa = 11.84FYY932 pKa = 11.49DD933 pKa = 3.74VVGTNLVEE941 pKa = 4.31IDD943 pKa = 3.83PGDD946 pKa = 3.59LAGLGISCSKK956 pKa = 10.48EE957 pKa = 3.72PNNNDD962 pKa = 3.49LGTGYY967 pKa = 9.33PAQSLDD973 pKa = 4.42NSYY976 pKa = 10.41LWPMDD981 pKa = 3.14TGVNPGYY988 pKa = 10.93LNGVWDD994 pKa = 4.14PAQGNLVADD1003 pKa = 4.51ADD1005 pKa = 5.0LICNMGLLLDD1015 pKa = 4.27GQSRR1019 pKa = 11.84AITYY1023 pKa = 8.9EE1024 pKa = 3.82LEE1026 pKa = 4.2ADD1028 pKa = 3.37TRR1030 pKa = 11.84GVYY1033 pKa = 10.26FNYY1036 pKa = 10.35AISRR1040 pKa = 11.84SRR1042 pKa = 11.84EE1043 pKa = 3.68HH1044 pKa = 7.16RR1045 pKa = 11.84EE1046 pKa = 3.88NGVDD1050 pKa = 3.89GPDD1053 pKa = 3.29VVPGNDD1059 pKa = 3.04VTRR1062 pKa = 11.84EE1063 pKa = 3.78RR1064 pKa = 11.84TTVRR1068 pKa = 11.84SVPNVGLSKK1077 pKa = 10.68VSSAEE1082 pKa = 3.82QVSLLEE1088 pKa = 4.39PFDD1091 pKa = 5.06FIITASNLHH1100 pKa = 5.8TDD1102 pKa = 3.34EE1103 pKa = 4.74LAYY1106 pKa = 10.49FPEE1109 pKa = 4.41VRR1111 pKa = 11.84DD1112 pKa = 3.69TLPAGMEE1119 pKa = 3.94LTAPPVLEE1127 pKa = 4.31SGAPVGSSCTGVAGDD1142 pKa = 4.37SDD1144 pKa = 5.1FICSLGDD1151 pKa = 4.02GIPPQAEE1158 pKa = 3.91VVIRR1162 pKa = 11.84VPVRR1166 pKa = 11.84VVAGGAQTLINEE1178 pKa = 4.67AFLHH1182 pKa = 6.73LDD1184 pKa = 3.13TDD1186 pKa = 4.23LEE1188 pKa = 4.3FDD1190 pKa = 4.25EE1191 pKa = 5.37EE1192 pKa = 4.34PPAAAEE1198 pKa = 4.51DD1199 pKa = 4.28DD1200 pKa = 4.13DD1201 pKa = 5.22DD1202 pKa = 4.57VVVVVSSLAGRR1213 pKa = 11.84VYY1215 pKa = 10.75HH1216 pKa = 7.55DD1217 pKa = 3.89NDD1219 pKa = 3.21RR1220 pKa = 11.84SGAYY1224 pKa = 8.93TSGNPPIAGVTLTLTGTSLAGDD1246 pKa = 4.44SITRR1250 pKa = 11.84TTVTEE1255 pKa = 3.77PDD1257 pKa = 3.04GTYY1260 pKa = 10.72LFEE1263 pKa = 4.4EE1264 pKa = 5.25LPAGSYY1270 pKa = 10.76SVTQTQPGGWIDD1282 pKa = 3.91GPVTQGSEE1290 pKa = 4.09GGSPSVNLVDD1300 pKa = 5.19NVQLPPDD1307 pKa = 4.06TAAVQYY1313 pKa = 11.32DD1314 pKa = 3.78FGEE1317 pKa = 4.28YY1318 pKa = 10.41LADD1321 pKa = 5.45DD1322 pKa = 4.72LDD1324 pKa = 4.21EE1325 pKa = 4.61LASISGHH1332 pKa = 5.74VYY1334 pKa = 10.57FDD1336 pKa = 3.93VNDD1339 pKa = 3.7NGQMNDD1345 pKa = 3.88SEE1347 pKa = 4.53PGIADD1352 pKa = 3.2VVVNLWRR1359 pKa = 11.84DD1360 pKa = 3.16GALIATQRR1368 pKa = 11.84TDD1370 pKa = 3.19DD1371 pKa = 4.08DD1372 pKa = 3.77GVYY1375 pKa = 10.26FFEE1378 pKa = 4.44NLAPGTYY1385 pKa = 9.01RR1386 pKa = 11.84VTEE1389 pKa = 4.11SQPAEE1394 pKa = 3.97WIDD1397 pKa = 3.3ARR1399 pKa = 11.84EE1400 pKa = 4.13SVGQGATSPGSSPQNDD1416 pKa = 3.24VFDD1419 pKa = 4.25GVEE1422 pKa = 3.89LLGGDD1427 pKa = 3.43VAINYY1432 pKa = 8.77NFGEE1436 pKa = 4.36LRR1438 pKa = 11.84NVPPIPTIGVYY1449 pKa = 11.11GLMLLVLMMTGLAQRR1464 pKa = 11.84KK1465 pKa = 8.74RR1466 pKa = 11.84RR1467 pKa = 11.84SLSS1470 pKa = 3.14
MM1 pKa = 7.21TAGFSGTRR9 pKa = 11.84EE10 pKa = 3.69RR11 pKa = 11.84GFSFFRR17 pKa = 11.84GLAQSVPGKK26 pKa = 10.57KK27 pKa = 10.26LLTSLLAAPMLMLVAGFASAQSVDD51 pKa = 3.51LVLNHH56 pKa = 6.41GQDD59 pKa = 3.61LPTINATDD67 pKa = 3.54TVTFTVTVDD76 pKa = 3.14NSDD79 pKa = 3.51PGFTADD85 pKa = 3.74AANVVLDD92 pKa = 3.89YY93 pKa = 10.97QISPTDD99 pKa = 3.46TLVSVTPSTGCSAPDD114 pKa = 3.25ASNAFSCNLGTLLHH128 pKa = 6.18GTDD131 pKa = 3.32TSLDD135 pKa = 3.2IVVRR139 pKa = 11.84GTGTGVSNFPYY150 pKa = 10.76VMQTTATVSTSNNDD164 pKa = 2.78IDD166 pKa = 4.19PSNNTEE172 pKa = 3.87QRR174 pKa = 11.84NVTVNAGTDD183 pKa = 3.29LGVVLNSSPGMTPSGGAWTHH203 pKa = 6.97DD204 pKa = 2.81IVVDD208 pKa = 3.6NYY210 pKa = 11.46GPIDD214 pKa = 3.54ATNAVIHH221 pKa = 6.92LDD223 pKa = 3.67LPVGVVVQSVPAGCSVAAPGVVCQVGALSVGDD255 pKa = 4.31SLPLGSIVTQVAVASGSNLSATALIDD281 pKa = 5.02SLDD284 pKa = 3.67QYY286 pKa = 11.47DD287 pKa = 4.14GNSINQQSTRR297 pKa = 11.84QVEE300 pKa = 4.39VTPGSDD306 pKa = 2.95LSVGLSHH313 pKa = 7.1TAGTILEE320 pKa = 4.3GQPFNLTASPAYY332 pKa = 9.0TGEE335 pKa = 4.21VPDD338 pKa = 4.66IPALVIDD345 pKa = 4.83VDD347 pKa = 4.05PGLQILSPANFSQNGWSCSVTGQQVSCSGLDD378 pKa = 3.35VGSMVPGSSQSLGDD392 pKa = 3.32VSVQVSGATAGSFAANQAQISADD415 pKa = 3.52YY416 pKa = 10.53SVKK419 pKa = 10.21PDD421 pKa = 3.33SDD423 pKa = 3.53LSNNVAGTTPVIEE436 pKa = 4.28SANLNVGINKK446 pKa = 9.64SPPNRR451 pKa = 11.84NLRR454 pKa = 11.84TIGAAYY460 pKa = 9.25PFDD463 pKa = 3.56YY464 pKa = 10.59QIRR467 pKa = 11.84VRR469 pKa = 11.84NNGNIAFWGDD479 pKa = 3.73LTWSDD484 pKa = 4.03NVPAGLTVTAINGPAGWSCSPALPVTGAATIDD516 pKa = 3.68CVRR519 pKa = 11.84TYY521 pKa = 11.12TEE523 pKa = 4.24GAPLAVNATQDD534 pKa = 3.68FTVTAYY540 pKa = 10.74ANGAISGNITNQACLTQIDD559 pKa = 4.38HH560 pKa = 7.74DD561 pKa = 4.8LAFLDD566 pKa = 4.13NVDD569 pKa = 4.19PAPCNGATINAQALNDD585 pKa = 3.98SADD588 pKa = 3.53LRR590 pKa = 11.84VVKK593 pKa = 9.0TASAATVNAGEE604 pKa = 4.2ALTYY608 pKa = 8.13TLRR611 pKa = 11.84IVNDD615 pKa = 4.59GPATASDD622 pKa = 3.72VDD624 pKa = 3.98LTDD627 pKa = 3.7AFSALFTGSAGNSFQGATVTNATASGGTCGTSGLAGNVGSRR668 pKa = 11.84TLSCNFASIPVCSGTDD684 pKa = 3.39CPSIEE689 pKa = 4.41VTVLPLGNTSSGVPLVRR706 pKa = 11.84NNTASAYY713 pKa = 10.39SNEE716 pKa = 4.47TADD719 pKa = 4.09PDD721 pKa = 3.89YY722 pKa = 11.21GNNPGSVSTTINPLTDD738 pKa = 3.87LLVSKK743 pKa = 10.68DD744 pKa = 3.42VTVWSGNLGTPLEE757 pKa = 4.11YY758 pKa = 10.24RR759 pKa = 11.84LQIEE763 pKa = 4.71NIGASGASGLSLEE776 pKa = 5.2DD777 pKa = 3.84VLPHH781 pKa = 7.03DD782 pKa = 4.6LTYY785 pKa = 11.31LSTDD789 pKa = 3.29PGASASCATEE799 pKa = 3.93PGANTTTTTGNDD811 pKa = 3.26EE812 pKa = 4.23IACTRR817 pKa = 11.84STLPRR822 pKa = 11.84NATWTVTVNTRR833 pKa = 11.84PNHH836 pKa = 6.47GIPAGTTLVNSVDD849 pKa = 3.96AEE851 pKa = 4.3TDD853 pKa = 3.38TPEE856 pKa = 4.11TDD858 pKa = 3.54LSNNADD864 pKa = 3.6TADD867 pKa = 3.83AEE869 pKa = 4.33VGEE872 pKa = 4.51AVVDD876 pKa = 4.01LAVIKK881 pKa = 10.65ADD883 pKa = 3.53TPDD886 pKa = 3.5PVFVGDD892 pKa = 3.43RR893 pKa = 11.84VTYY896 pKa = 8.78SLRR899 pKa = 11.84IINNGPSVATDD910 pKa = 3.18ATIYY914 pKa = 10.67DD915 pKa = 4.32YY916 pKa = 11.37LPSAGLRR923 pKa = 11.84YY924 pKa = 9.89VEE926 pKa = 4.0NSIRR930 pKa = 11.84FYY932 pKa = 11.49DD933 pKa = 3.74VVGTNLVEE941 pKa = 4.31IDD943 pKa = 3.83PGDD946 pKa = 3.59LAGLGISCSKK956 pKa = 10.48EE957 pKa = 3.72PNNNDD962 pKa = 3.49LGTGYY967 pKa = 9.33PAQSLDD973 pKa = 4.42NSYY976 pKa = 10.41LWPMDD981 pKa = 3.14TGVNPGYY988 pKa = 10.93LNGVWDD994 pKa = 4.14PAQGNLVADD1003 pKa = 4.51ADD1005 pKa = 5.0LICNMGLLLDD1015 pKa = 4.27GQSRR1019 pKa = 11.84AITYY1023 pKa = 8.9EE1024 pKa = 3.82LEE1026 pKa = 4.2ADD1028 pKa = 3.37TRR1030 pKa = 11.84GVYY1033 pKa = 10.26FNYY1036 pKa = 10.35AISRR1040 pKa = 11.84SRR1042 pKa = 11.84EE1043 pKa = 3.68HH1044 pKa = 7.16RR1045 pKa = 11.84EE1046 pKa = 3.88NGVDD1050 pKa = 3.89GPDD1053 pKa = 3.29VVPGNDD1059 pKa = 3.04VTRR1062 pKa = 11.84EE1063 pKa = 3.78RR1064 pKa = 11.84TTVRR1068 pKa = 11.84SVPNVGLSKK1077 pKa = 10.68VSSAEE1082 pKa = 3.82QVSLLEE1088 pKa = 4.39PFDD1091 pKa = 5.06FIITASNLHH1100 pKa = 5.8TDD1102 pKa = 3.34EE1103 pKa = 4.74LAYY1106 pKa = 10.49FPEE1109 pKa = 4.41VRR1111 pKa = 11.84DD1112 pKa = 3.69TLPAGMEE1119 pKa = 3.94LTAPPVLEE1127 pKa = 4.31SGAPVGSSCTGVAGDD1142 pKa = 4.37SDD1144 pKa = 5.1FICSLGDD1151 pKa = 4.02GIPPQAEE1158 pKa = 3.91VVIRR1162 pKa = 11.84VPVRR1166 pKa = 11.84VVAGGAQTLINEE1178 pKa = 4.67AFLHH1182 pKa = 6.73LDD1184 pKa = 3.13TDD1186 pKa = 4.23LEE1188 pKa = 4.3FDD1190 pKa = 4.25EE1191 pKa = 5.37EE1192 pKa = 4.34PPAAAEE1198 pKa = 4.51DD1199 pKa = 4.28DD1200 pKa = 4.13DD1201 pKa = 5.22DD1202 pKa = 4.57VVVVVSSLAGRR1213 pKa = 11.84VYY1215 pKa = 10.75HH1216 pKa = 7.55DD1217 pKa = 3.89NDD1219 pKa = 3.21RR1220 pKa = 11.84SGAYY1224 pKa = 8.93TSGNPPIAGVTLTLTGTSLAGDD1246 pKa = 4.44SITRR1250 pKa = 11.84TTVTEE1255 pKa = 3.77PDD1257 pKa = 3.04GTYY1260 pKa = 10.72LFEE1263 pKa = 4.4EE1264 pKa = 5.25LPAGSYY1270 pKa = 10.76SVTQTQPGGWIDD1282 pKa = 3.91GPVTQGSEE1290 pKa = 4.09GGSPSVNLVDD1300 pKa = 5.19NVQLPPDD1307 pKa = 4.06TAAVQYY1313 pKa = 11.32DD1314 pKa = 3.78FGEE1317 pKa = 4.28YY1318 pKa = 10.41LADD1321 pKa = 5.45DD1322 pKa = 4.72LDD1324 pKa = 4.21EE1325 pKa = 4.61LASISGHH1332 pKa = 5.74VYY1334 pKa = 10.57FDD1336 pKa = 3.93VNDD1339 pKa = 3.7NGQMNDD1345 pKa = 3.88SEE1347 pKa = 4.53PGIADD1352 pKa = 3.2VVVNLWRR1359 pKa = 11.84DD1360 pKa = 3.16GALIATQRR1368 pKa = 11.84TDD1370 pKa = 3.19DD1371 pKa = 4.08DD1372 pKa = 3.77GVYY1375 pKa = 10.26FFEE1378 pKa = 4.44NLAPGTYY1385 pKa = 9.01RR1386 pKa = 11.84VTEE1389 pKa = 4.11SQPAEE1394 pKa = 3.97WIDD1397 pKa = 3.3ARR1399 pKa = 11.84EE1400 pKa = 4.13SVGQGATSPGSSPQNDD1416 pKa = 3.24VFDD1419 pKa = 4.25GVEE1422 pKa = 3.89LLGGDD1427 pKa = 3.43VAINYY1432 pKa = 8.77NFGEE1436 pKa = 4.36LRR1438 pKa = 11.84NVPPIPTIGVYY1449 pKa = 11.11GLMLLVLMMTGLAQRR1464 pKa = 11.84KK1465 pKa = 8.74RR1466 pKa = 11.84RR1467 pKa = 11.84SLSS1470 pKa = 3.14
Molecular weight: 152.18 kDa
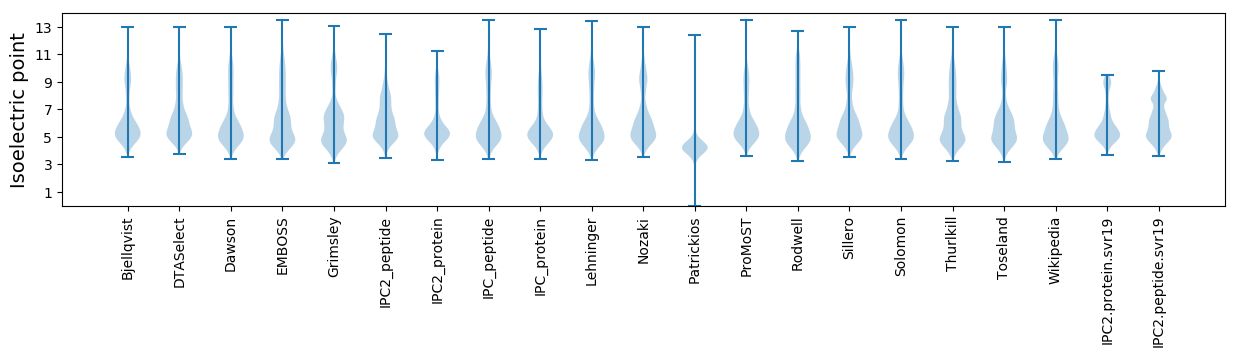
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H2HNW5|A0A1H2HNW5_9PSED ATP synthase gamma chain OS=Pseudomonas salegens OX=1434072 GN=atpG PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.46RR12 pKa = 11.84KK13 pKa = 9.37RR14 pKa = 11.84NHH16 pKa = 5.37GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.3NGRR28 pKa = 11.84AVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.93GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.46RR12 pKa = 11.84KK13 pKa = 9.37RR14 pKa = 11.84NHH16 pKa = 5.37GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.3NGRR28 pKa = 11.84AVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.93GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1136679 |
29 |
2852 |
332.9 |
36.67 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.564 ± 0.043 | 1.038 ± 0.013 |
5.765 ± 0.037 | 5.869 ± 0.035 |
3.555 ± 0.028 | 7.662 ± 0.036 |
2.349 ± 0.02 | 4.947 ± 0.032 |
2.931 ± 0.039 | 11.798 ± 0.061 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.467 ± 0.021 | 3.112 ± 0.028 |
4.848 ± 0.028 | 5.009 ± 0.041 |
6.519 ± 0.044 | 5.9 ± 0.033 |
4.832 ± 0.028 | 6.837 ± 0.034 |
1.482 ± 0.021 | 2.516 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
