
Scytonema hofmannii PCC 7110
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Cyanobacteria/Melainabacteria group; Cyanobacteria; Nostocales; Scytonemataceae; Scytonema; Scytonema hofmannii
Average proteome isoelectric point is 6.38
Get precalculated fractions of proteins
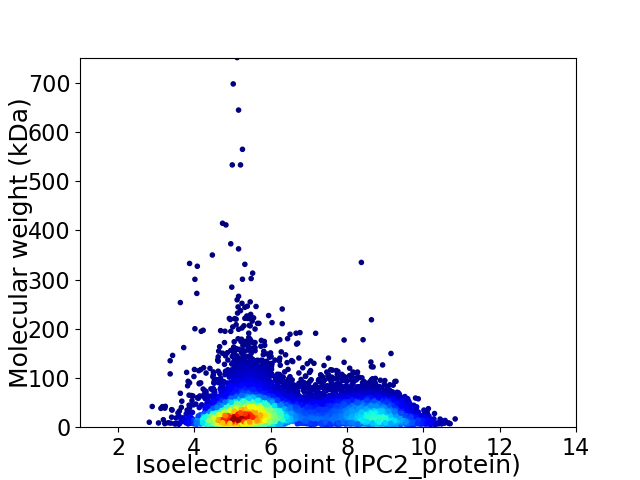
Virtual 2D-PAGE plot for 9705 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A139WWM3|A0A139WWM3_9CYAN Uncharacterized protein OS=Scytonema hofmannii PCC 7110 OX=128403 GN=WA1_44735 PE=4 SV=1
MM1 pKa = 7.32AHH3 pKa = 7.55DD4 pKa = 4.41GTTTLPTQYY13 pKa = 9.13NTSAEE18 pKa = 4.14NLTEE22 pKa = 4.04EE23 pKa = 4.52DD24 pKa = 3.66TLVANSEE31 pKa = 4.02MPMQYY36 pKa = 8.57NTSAEE41 pKa = 4.1NLTVEE46 pKa = 4.52DD47 pKa = 3.82TLVANSEE54 pKa = 4.02MPMQYY59 pKa = 8.57NTSAEE64 pKa = 4.1NLTAEE69 pKa = 4.52DD70 pKa = 4.05TLVGNPEE77 pKa = 4.24EE78 pKa = 4.4PVVLPDD84 pKa = 3.88INRR87 pKa = 11.84PGYY90 pKa = 9.35WLVGDD95 pKa = 4.34HH96 pKa = 6.36ATFVADD102 pKa = 4.93GEE104 pKa = 4.63DD105 pKa = 3.64TNGQYY110 pKa = 11.33SLFDD114 pKa = 4.89FYY116 pKa = 10.98TLPQGGPFPHH126 pKa = 6.76IHH128 pKa = 5.1RR129 pKa = 11.84TEE131 pKa = 3.88NEE133 pKa = 3.34FAYY136 pKa = 9.9ILDD139 pKa = 4.15GEE141 pKa = 4.51ISYY144 pKa = 10.6QLNDD148 pKa = 3.61EE149 pKa = 5.03VITATPGTFVYY160 pKa = 10.09KK161 pKa = 10.49HH162 pKa = 6.19QDD164 pKa = 3.84DD165 pKa = 3.18IHH167 pKa = 7.89GFVNLGDD174 pKa = 3.75TPSRR178 pKa = 11.84HH179 pKa = 6.76LEE181 pKa = 3.88FTLPPGLEE189 pKa = 3.85NAFATVGVPGSILEE203 pKa = 4.21PPPNEE208 pKa = 4.03IPPSEE213 pKa = 4.04ILDD216 pKa = 3.84NYY218 pKa = 9.79TEE220 pKa = 4.05VLAEE224 pKa = 4.16YY225 pKa = 10.35GVEE228 pKa = 4.01AQNSIIFPSAGLNEE242 pKa = 4.46TLTGVPEE249 pKa = 4.24VTLLRR254 pKa = 11.84PGQADD259 pKa = 3.61TAVSATLALSNGTAIPVDD277 pKa = 4.62FAAGEE282 pKa = 4.04RR283 pKa = 11.84TKK285 pKa = 10.53TVEE288 pKa = 3.77IPVDD292 pKa = 3.74GSEE295 pKa = 4.65VPGQTLEE302 pKa = 4.28LAITNPTDD310 pKa = 3.45GAIVGLLQDD319 pKa = 4.07EE320 pKa = 4.65LVLTFGEE327 pKa = 4.58DD328 pKa = 2.97NTYY331 pKa = 9.13TLEE334 pKa = 4.56NGDD337 pKa = 4.7KK338 pKa = 9.98PDD340 pKa = 3.73SFPTILPNDD349 pKa = 3.67EE350 pKa = 4.16EE351 pKa = 4.19RR352 pKa = 11.84LKK354 pKa = 11.33VSLGNDD360 pKa = 3.08EE361 pKa = 4.39YY362 pKa = 11.49TFVATAADD370 pKa = 3.82TDD372 pKa = 4.27GKK374 pKa = 10.76ISLLDD379 pKa = 3.36VSVRR383 pKa = 11.84PGTQNEE389 pKa = 4.49SLLSAPTDD397 pKa = 3.45LGFYY401 pKa = 10.34VVDD404 pKa = 4.01GNVTFEE410 pKa = 4.92LGDD413 pKa = 3.6EE414 pKa = 4.52SFTAAQDD421 pKa = 3.55TFVFLPEE428 pKa = 4.44GNSFALTNQGTLPARR443 pKa = 11.84TLAFSTSPEE452 pKa = 3.66IEE454 pKa = 4.19EE455 pKa = 4.99YY456 pKa = 9.31IASMGIGDD464 pKa = 4.1TQSNSPLSIMEE475 pKa = 4.45PPLTSDD481 pKa = 3.86LSTMISTEE489 pKa = 4.0SLVV492 pKa = 3.28
MM1 pKa = 7.32AHH3 pKa = 7.55DD4 pKa = 4.41GTTTLPTQYY13 pKa = 9.13NTSAEE18 pKa = 4.14NLTEE22 pKa = 4.04EE23 pKa = 4.52DD24 pKa = 3.66TLVANSEE31 pKa = 4.02MPMQYY36 pKa = 8.57NTSAEE41 pKa = 4.1NLTVEE46 pKa = 4.52DD47 pKa = 3.82TLVANSEE54 pKa = 4.02MPMQYY59 pKa = 8.57NTSAEE64 pKa = 4.1NLTAEE69 pKa = 4.52DD70 pKa = 4.05TLVGNPEE77 pKa = 4.24EE78 pKa = 4.4PVVLPDD84 pKa = 3.88INRR87 pKa = 11.84PGYY90 pKa = 9.35WLVGDD95 pKa = 4.34HH96 pKa = 6.36ATFVADD102 pKa = 4.93GEE104 pKa = 4.63DD105 pKa = 3.64TNGQYY110 pKa = 11.33SLFDD114 pKa = 4.89FYY116 pKa = 10.98TLPQGGPFPHH126 pKa = 6.76IHH128 pKa = 5.1RR129 pKa = 11.84TEE131 pKa = 3.88NEE133 pKa = 3.34FAYY136 pKa = 9.9ILDD139 pKa = 4.15GEE141 pKa = 4.51ISYY144 pKa = 10.6QLNDD148 pKa = 3.61EE149 pKa = 5.03VITATPGTFVYY160 pKa = 10.09KK161 pKa = 10.49HH162 pKa = 6.19QDD164 pKa = 3.84DD165 pKa = 3.18IHH167 pKa = 7.89GFVNLGDD174 pKa = 3.75TPSRR178 pKa = 11.84HH179 pKa = 6.76LEE181 pKa = 3.88FTLPPGLEE189 pKa = 3.85NAFATVGVPGSILEE203 pKa = 4.21PPPNEE208 pKa = 4.03IPPSEE213 pKa = 4.04ILDD216 pKa = 3.84NYY218 pKa = 9.79TEE220 pKa = 4.05VLAEE224 pKa = 4.16YY225 pKa = 10.35GVEE228 pKa = 4.01AQNSIIFPSAGLNEE242 pKa = 4.46TLTGVPEE249 pKa = 4.24VTLLRR254 pKa = 11.84PGQADD259 pKa = 3.61TAVSATLALSNGTAIPVDD277 pKa = 4.62FAAGEE282 pKa = 4.04RR283 pKa = 11.84TKK285 pKa = 10.53TVEE288 pKa = 3.77IPVDD292 pKa = 3.74GSEE295 pKa = 4.65VPGQTLEE302 pKa = 4.28LAITNPTDD310 pKa = 3.45GAIVGLLQDD319 pKa = 4.07EE320 pKa = 4.65LVLTFGEE327 pKa = 4.58DD328 pKa = 2.97NTYY331 pKa = 9.13TLEE334 pKa = 4.56NGDD337 pKa = 4.7KK338 pKa = 9.98PDD340 pKa = 3.73SFPTILPNDD349 pKa = 3.67EE350 pKa = 4.16EE351 pKa = 4.19RR352 pKa = 11.84LKK354 pKa = 11.33VSLGNDD360 pKa = 3.08EE361 pKa = 4.39YY362 pKa = 11.49TFVATAADD370 pKa = 3.82TDD372 pKa = 4.27GKK374 pKa = 10.76ISLLDD379 pKa = 3.36VSVRR383 pKa = 11.84PGTQNEE389 pKa = 4.49SLLSAPTDD397 pKa = 3.45LGFYY401 pKa = 10.34VVDD404 pKa = 4.01GNVTFEE410 pKa = 4.92LGDD413 pKa = 3.6EE414 pKa = 4.52SFTAAQDD421 pKa = 3.55TFVFLPEE428 pKa = 4.44GNSFALTNQGTLPARR443 pKa = 11.84TLAFSTSPEE452 pKa = 3.66IEE454 pKa = 4.19EE455 pKa = 4.99YY456 pKa = 9.31IASMGIGDD464 pKa = 4.1TQSNSPLSIMEE475 pKa = 4.45PPLTSDD481 pKa = 3.86LSTMISTEE489 pKa = 4.0SLVV492 pKa = 3.28
Molecular weight: 52.85 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A139WSV5|A0A139WSV5_9CYAN Uncharacterized protein OS=Scytonema hofmannii PCC 7110 OX=128403 GN=WA1_06785 PE=4 SV=1
MM1 pKa = 7.17NFKK4 pKa = 10.68VLLCSSVITINALAILPVYY23 pKa = 9.74AQQLTPQQAQIARR36 pKa = 11.84EE37 pKa = 3.84RR38 pKa = 11.84LRR40 pKa = 11.84CIMLGQGGDD49 pKa = 3.42LSEE52 pKa = 4.49PLSGEE57 pKa = 3.92LQYY60 pKa = 11.45LQLRR64 pKa = 11.84RR65 pKa = 11.84LNPQLADD72 pKa = 3.46RR73 pKa = 11.84CRR75 pKa = 11.84NLLKK79 pKa = 10.86GIFPGQYY86 pKa = 9.46LPNRR90 pKa = 11.84TRR92 pKa = 11.84RR93 pKa = 11.84PVVNPFPPGSLIKK106 pKa = 9.82EE107 pKa = 4.14QNVPLINSTPATRR120 pKa = 3.99
MM1 pKa = 7.17NFKK4 pKa = 10.68VLLCSSVITINALAILPVYY23 pKa = 9.74AQQLTPQQAQIARR36 pKa = 11.84EE37 pKa = 3.84RR38 pKa = 11.84LRR40 pKa = 11.84CIMLGQGGDD49 pKa = 3.42LSEE52 pKa = 4.49PLSGEE57 pKa = 3.92LQYY60 pKa = 11.45LQLRR64 pKa = 11.84RR65 pKa = 11.84LNPQLADD72 pKa = 3.46RR73 pKa = 11.84CRR75 pKa = 11.84NLLKK79 pKa = 10.86GIFPGQYY86 pKa = 9.46LPNRR90 pKa = 11.84TRR92 pKa = 11.84RR93 pKa = 11.84PVVNPFPPGSLIKK106 pKa = 9.82EE107 pKa = 4.14QNVPLINSTPATRR120 pKa = 3.99
Molecular weight: 13.43 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3138299 |
31 |
6702 |
323.4 |
36.15 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.684 ± 0.029 | 0.994 ± 0.008 |
4.854 ± 0.019 | 6.496 ± 0.026 |
4.023 ± 0.017 | 6.466 ± 0.033 |
1.811 ± 0.013 | 6.624 ± 0.024 |
5.112 ± 0.027 | 10.925 ± 0.031 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.748 ± 0.01 | 4.564 ± 0.023 |
4.517 ± 0.022 | 5.318 ± 0.028 |
5.19 ± 0.021 | 6.722 ± 0.023 |
5.773 ± 0.022 | 6.692 ± 0.019 |
1.412 ± 0.01 | 3.076 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
