
Wenzhou shrimp virus 9
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.45
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A1L3KEL7|A0A1L3KEL7_9VIRU RNA-directed RNA polymerase OS=Wenzhou shrimp virus 9 OX=1923656 PE=4 SV=1
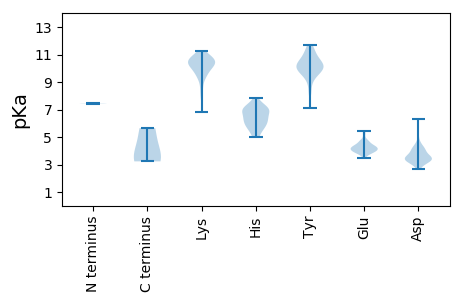
MM1 pKa = 7.45TNQHH5 pKa = 5.69VCPICKK11 pKa = 9.73RR12 pKa = 11.84SFRR15 pKa = 11.84TAQGLADD22 pKa = 3.93HH23 pKa = 6.69KK24 pKa = 10.55RR25 pKa = 11.84DD26 pKa = 3.36RR27 pKa = 11.84HH28 pKa = 5.03EE29 pKa = 4.37ACSTGPAPKK38 pKa = 9.18PQRR41 pKa = 11.84VRR43 pKa = 11.84PKK45 pKa = 10.22RR46 pKa = 11.84EE47 pKa = 3.54QRR49 pKa = 11.84QAAPVSVPTSSGPARR64 pKa = 11.84YY65 pKa = 9.6SNRR68 pKa = 11.84VTLQGSNEE76 pKa = 3.94RR77 pKa = 11.84FRR79 pKa = 11.84TDD81 pKa = 3.77DD82 pKa = 3.6YY83 pKa = 11.04PASRR87 pKa = 11.84LKK89 pKa = 10.19TGTLLTTIPVSPDD102 pKa = 2.98LVPRR106 pKa = 11.84LSTQAKK112 pKa = 7.82AYY114 pKa = 10.37QRR116 pKa = 11.84IKK118 pKa = 8.15YY119 pKa = 9.74HH120 pKa = 6.24SVALHH125 pKa = 6.41IDD127 pKa = 3.59AKK129 pKa = 11.09GSTAQSGGYY138 pKa = 9.34LAVFISDD145 pKa = 3.23ITDD148 pKa = 3.32EE149 pKa = 4.73EE150 pKa = 4.49ISLEE154 pKa = 4.1RR155 pKa = 11.84AGSFGGSVGKK165 pKa = 10.49KK166 pKa = 6.83YY167 pKa = 10.12WEE169 pKa = 4.3NATVTATNLTPLFYY183 pKa = 10.15TSRR186 pKa = 11.84GEE188 pKa = 5.45DD189 pKa = 3.25EE190 pKa = 5.31RR191 pKa = 11.84LWSPGYY197 pKa = 9.77FAIYY201 pKa = 10.12CDD203 pKa = 4.06GSSNTAISLTYY214 pKa = 10.3RR215 pKa = 11.84LTWSVTLSVPSSEE228 pKa = 5.12LKK230 pKa = 10.03QADD233 pKa = 4.43VITTMVDD240 pKa = 3.08LWPTAGKK247 pKa = 10.6ASFSDD252 pKa = 3.38KK253 pKa = 11.09DD254 pKa = 3.61GSEE257 pKa = 4.2EE258 pKa = 4.06NLFEE262 pKa = 4.8PSLPVGTVVRR272 pKa = 11.84FPYY275 pKa = 10.57AVGIEE280 pKa = 4.18YY281 pKa = 10.58KK282 pKa = 10.48EE283 pKa = 4.21GAGDD287 pKa = 3.63TGTVPIMFAVVTASSGLKK305 pKa = 10.2ASNDD309 pKa = 3.55RR310 pKa = 11.84VDD312 pKa = 4.05TSVVWQSDD320 pKa = 3.34VSRR323 pKa = 11.84RR324 pKa = 11.84LLYY327 pKa = 9.88PKK329 pKa = 10.06SIRR332 pKa = 11.84LIVEE336 pKa = 3.78KK337 pKa = 10.71RR338 pKa = 11.84PGEE341 pKa = 4.02VSRR344 pKa = 11.84AGQVSQFSPCPPTSLASSEE363 pKa = 4.43TPSTEE368 pKa = 3.82TVGNNEE374 pKa = 3.76NRR376 pKa = 11.84PTCSVRR382 pKa = 11.84FTTPSSSDD390 pKa = 3.16LMKK393 pKa = 10.96SMHH396 pKa = 5.96GCKK399 pKa = 9.58TSMSDD404 pKa = 2.94LRR406 pKa = 11.84ARR408 pKa = 11.84PVCTVSSTLPPSPNPSQEE426 pKa = 4.42SILLTFLEE434 pKa = 4.64SQVPRR439 pKa = 11.84YY440 pKa = 8.27QAARR444 pKa = 11.84IVSQEE449 pKa = 4.06VKK451 pKa = 8.29THH453 pKa = 6.18EE454 pKa = 4.43FPLLHH459 pKa = 6.52HH460 pKa = 6.91RR461 pKa = 11.84NHH463 pKa = 6.65LRR465 pKa = 11.84LEE467 pKa = 4.22MHH469 pKa = 6.99WDD471 pKa = 4.15LVPLMRR477 pKa = 11.84GRR479 pKa = 11.84RR480 pKa = 11.84IPVYY484 pKa = 9.57EE485 pKa = 4.4LSRR488 pKa = 11.84HH489 pKa = 5.57FDD491 pKa = 3.51DD492 pKa = 6.33DD493 pKa = 4.22FSLKK497 pKa = 9.41FTINNTVSFRR507 pKa = 11.84EE508 pKa = 3.86VVEE511 pKa = 3.94AVEE514 pKa = 3.87VLNRR518 pKa = 11.84GRR520 pKa = 11.84YY521 pKa = 8.22FCLGFIALCLQHH533 pKa = 6.89HH534 pKa = 6.95FGADD538 pKa = 3.16SSEE541 pKa = 4.21VVPEE545 pKa = 4.0EE546 pKa = 4.68FPSVWYY552 pKa = 10.07DD553 pKa = 3.26EE554 pKa = 4.49SFHH557 pKa = 6.49YY558 pKa = 10.38RR559 pKa = 11.84LPCRR563 pKa = 11.84SCHH566 pKa = 7.01DD567 pKa = 3.8NMEE570 pKa = 4.3SDD572 pKa = 3.96YY573 pKa = 11.47SFEE576 pKa = 4.03EE577 pKa = 3.99LDD579 pKa = 4.09PEE581 pKa = 4.52VEE583 pKa = 4.05YY584 pKa = 11.04DD585 pKa = 3.23
MM1 pKa = 7.45TNQHH5 pKa = 5.69VCPICKK11 pKa = 9.73RR12 pKa = 11.84SFRR15 pKa = 11.84TAQGLADD22 pKa = 3.93HH23 pKa = 6.69KK24 pKa = 10.55RR25 pKa = 11.84DD26 pKa = 3.36RR27 pKa = 11.84HH28 pKa = 5.03EE29 pKa = 4.37ACSTGPAPKK38 pKa = 9.18PQRR41 pKa = 11.84VRR43 pKa = 11.84PKK45 pKa = 10.22RR46 pKa = 11.84EE47 pKa = 3.54QRR49 pKa = 11.84QAAPVSVPTSSGPARR64 pKa = 11.84YY65 pKa = 9.6SNRR68 pKa = 11.84VTLQGSNEE76 pKa = 3.94RR77 pKa = 11.84FRR79 pKa = 11.84TDD81 pKa = 3.77DD82 pKa = 3.6YY83 pKa = 11.04PASRR87 pKa = 11.84LKK89 pKa = 10.19TGTLLTTIPVSPDD102 pKa = 2.98LVPRR106 pKa = 11.84LSTQAKK112 pKa = 7.82AYY114 pKa = 10.37QRR116 pKa = 11.84IKK118 pKa = 8.15YY119 pKa = 9.74HH120 pKa = 6.24SVALHH125 pKa = 6.41IDD127 pKa = 3.59AKK129 pKa = 11.09GSTAQSGGYY138 pKa = 9.34LAVFISDD145 pKa = 3.23ITDD148 pKa = 3.32EE149 pKa = 4.73EE150 pKa = 4.49ISLEE154 pKa = 4.1RR155 pKa = 11.84AGSFGGSVGKK165 pKa = 10.49KK166 pKa = 6.83YY167 pKa = 10.12WEE169 pKa = 4.3NATVTATNLTPLFYY183 pKa = 10.15TSRR186 pKa = 11.84GEE188 pKa = 5.45DD189 pKa = 3.25EE190 pKa = 5.31RR191 pKa = 11.84LWSPGYY197 pKa = 9.77FAIYY201 pKa = 10.12CDD203 pKa = 4.06GSSNTAISLTYY214 pKa = 10.3RR215 pKa = 11.84LTWSVTLSVPSSEE228 pKa = 5.12LKK230 pKa = 10.03QADD233 pKa = 4.43VITTMVDD240 pKa = 3.08LWPTAGKK247 pKa = 10.6ASFSDD252 pKa = 3.38KK253 pKa = 11.09DD254 pKa = 3.61GSEE257 pKa = 4.2EE258 pKa = 4.06NLFEE262 pKa = 4.8PSLPVGTVVRR272 pKa = 11.84FPYY275 pKa = 10.57AVGIEE280 pKa = 4.18YY281 pKa = 10.58KK282 pKa = 10.48EE283 pKa = 4.21GAGDD287 pKa = 3.63TGTVPIMFAVVTASSGLKK305 pKa = 10.2ASNDD309 pKa = 3.55RR310 pKa = 11.84VDD312 pKa = 4.05TSVVWQSDD320 pKa = 3.34VSRR323 pKa = 11.84RR324 pKa = 11.84LLYY327 pKa = 9.88PKK329 pKa = 10.06SIRR332 pKa = 11.84LIVEE336 pKa = 3.78KK337 pKa = 10.71RR338 pKa = 11.84PGEE341 pKa = 4.02VSRR344 pKa = 11.84AGQVSQFSPCPPTSLASSEE363 pKa = 4.43TPSTEE368 pKa = 3.82TVGNNEE374 pKa = 3.76NRR376 pKa = 11.84PTCSVRR382 pKa = 11.84FTTPSSSDD390 pKa = 3.16LMKK393 pKa = 10.96SMHH396 pKa = 5.96GCKK399 pKa = 9.58TSMSDD404 pKa = 2.94LRR406 pKa = 11.84ARR408 pKa = 11.84PVCTVSSTLPPSPNPSQEE426 pKa = 4.42SILLTFLEE434 pKa = 4.64SQVPRR439 pKa = 11.84YY440 pKa = 8.27QAARR444 pKa = 11.84IVSQEE449 pKa = 4.06VKK451 pKa = 8.29THH453 pKa = 6.18EE454 pKa = 4.43FPLLHH459 pKa = 6.52HH460 pKa = 6.91RR461 pKa = 11.84NHH463 pKa = 6.65LRR465 pKa = 11.84LEE467 pKa = 4.22MHH469 pKa = 6.99WDD471 pKa = 4.15LVPLMRR477 pKa = 11.84GRR479 pKa = 11.84RR480 pKa = 11.84IPVYY484 pKa = 9.57EE485 pKa = 4.4LSRR488 pKa = 11.84HH489 pKa = 5.57FDD491 pKa = 3.51DD492 pKa = 6.33DD493 pKa = 4.22FSLKK497 pKa = 9.41FTINNTVSFRR507 pKa = 11.84EE508 pKa = 3.86VVEE511 pKa = 3.94AVEE514 pKa = 3.87VLNRR518 pKa = 11.84GRR520 pKa = 11.84YY521 pKa = 8.22FCLGFIALCLQHH533 pKa = 6.89HH534 pKa = 6.95FGADD538 pKa = 3.16SSEE541 pKa = 4.21VVPEE545 pKa = 4.0EE546 pKa = 4.68FPSVWYY552 pKa = 10.07DD553 pKa = 3.26EE554 pKa = 4.49SFHH557 pKa = 6.49YY558 pKa = 10.38RR559 pKa = 11.84LPCRR563 pKa = 11.84SCHH566 pKa = 7.01DD567 pKa = 3.8NMEE570 pKa = 4.3SDD572 pKa = 3.96YY573 pKa = 11.47SFEE576 pKa = 4.03EE577 pKa = 3.99LDD579 pKa = 4.09PEE581 pKa = 4.52VEE583 pKa = 4.05YY584 pKa = 11.04DD585 pKa = 3.23
Molecular weight: 65.25 kDa
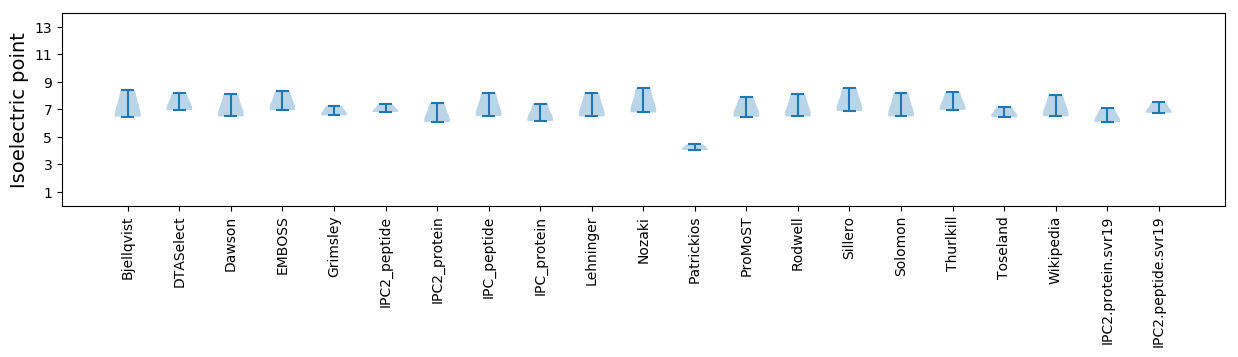
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KEL2|A0A1L3KEL2_9VIRU Putative capsid protein OS=Wenzhou shrimp virus 9 OX=1923656 PE=4 SV=1
MM1 pKa = 7.43GVFKK5 pKa = 10.82KK6 pKa = 10.48ILLKK10 pKa = 9.88TIKK13 pKa = 10.03EE14 pKa = 4.13VLEE17 pKa = 4.07EE18 pKa = 4.46EE19 pKa = 4.3EE20 pKa = 4.61LSTRR24 pKa = 11.84KK25 pKa = 9.89NPLTIGNVGKK35 pKa = 10.33SWLAAVGTYY44 pKa = 9.75KK45 pKa = 10.52KK46 pKa = 9.83HH47 pKa = 5.63LVVGALVGAGALAYY61 pKa = 10.0RR62 pKa = 11.84YY63 pKa = 7.13WKK65 pKa = 9.62NRR67 pKa = 11.84KK68 pKa = 8.68RR69 pKa = 11.84PSRR72 pKa = 11.84VVRR75 pKa = 11.84DD76 pKa = 3.58SFFRR80 pKa = 11.84AAITGDD86 pKa = 3.59VVPEE90 pKa = 4.2SKK92 pKa = 10.64VAGSPEE98 pKa = 4.09VNLLAPNSQCRR109 pKa = 11.84ILLKK113 pKa = 10.88NSDD116 pKa = 4.23GDD118 pKa = 3.89WVLTGCAVRR127 pKa = 11.84VDD129 pKa = 4.3LPIGPFLVFPQHH141 pKa = 6.02LCAGEE146 pKa = 3.91LAAMSSRR153 pKa = 11.84DD154 pKa = 3.49EE155 pKa = 4.42VIPLSLDD162 pKa = 3.1GLEE165 pKa = 5.16PIAADD170 pKa = 5.11LSMLPCPSKK179 pKa = 10.37LTSTGLASPTIGVVRR194 pKa = 11.84PGQRR198 pKa = 11.84CVVSVVGVDD207 pKa = 3.66GLGTTGPLQTSPVFGRR223 pKa = 11.84LIYY226 pKa = 10.5GATTNTGYY234 pKa = 10.71SGAAYY239 pKa = 10.04ASARR243 pKa = 11.84TVYY246 pKa = 10.17GVHH249 pKa = 6.18TNGGKK254 pKa = 10.53LNGGFALSYY263 pKa = 9.72VYY265 pKa = 11.68AMMKK269 pKa = 9.26VLKK272 pKa = 10.03GVRR275 pKa = 11.84DD276 pKa = 3.64EE277 pKa = 5.43SSEE280 pKa = 3.96NWIHH284 pKa = 7.15SVMADD289 pKa = 2.74KK290 pKa = 10.18STRR293 pKa = 11.84FLEE296 pKa = 3.89MDD298 pKa = 3.68YY299 pKa = 11.35EE300 pKa = 4.22RR301 pKa = 11.84VADD304 pKa = 4.3SIVMRR309 pKa = 11.84GDD311 pKa = 3.11DD312 pKa = 3.96GFYY315 pKa = 10.53HH316 pKa = 7.51AISGPAAEE324 pKa = 4.67GLVKK328 pKa = 10.76KK329 pKa = 10.38LDD331 pKa = 3.48KK332 pKa = 10.84YY333 pKa = 11.12KK334 pKa = 10.77YY335 pKa = 9.2DD336 pKa = 3.23AGMVDD341 pKa = 4.69FRR343 pKa = 11.84SKK345 pKa = 9.61TGHH348 pKa = 5.12TWSDD352 pKa = 2.68IDD354 pKa = 3.9PNHH357 pKa = 6.95WEE359 pKa = 4.08SANPEE364 pKa = 4.16TAQVLASADD373 pKa = 3.59TVPQGFHH380 pKa = 7.29LDD382 pKa = 3.48PSSRR386 pKa = 11.84EE387 pKa = 3.67SPYY390 pKa = 10.25IEE392 pKa = 4.4EE393 pKa = 4.29LASQIRR399 pKa = 11.84MFEE402 pKa = 4.17DD403 pKa = 3.3RR404 pKa = 11.84LKK406 pKa = 11.23SSLVDD411 pKa = 3.29TALKK415 pKa = 9.24KK416 pKa = 10.66QIMDD420 pKa = 4.21TFQQTLEE427 pKa = 4.13PSRR430 pKa = 11.84SLTLSLCRR438 pKa = 11.84PEE440 pKa = 4.27LQHH443 pKa = 7.16RR444 pKa = 11.84LAKK447 pKa = 10.81VMFGLHH453 pKa = 5.64
MM1 pKa = 7.43GVFKK5 pKa = 10.82KK6 pKa = 10.48ILLKK10 pKa = 9.88TIKK13 pKa = 10.03EE14 pKa = 4.13VLEE17 pKa = 4.07EE18 pKa = 4.46EE19 pKa = 4.3EE20 pKa = 4.61LSTRR24 pKa = 11.84KK25 pKa = 9.89NPLTIGNVGKK35 pKa = 10.33SWLAAVGTYY44 pKa = 9.75KK45 pKa = 10.52KK46 pKa = 9.83HH47 pKa = 5.63LVVGALVGAGALAYY61 pKa = 10.0RR62 pKa = 11.84YY63 pKa = 7.13WKK65 pKa = 9.62NRR67 pKa = 11.84KK68 pKa = 8.68RR69 pKa = 11.84PSRR72 pKa = 11.84VVRR75 pKa = 11.84DD76 pKa = 3.58SFFRR80 pKa = 11.84AAITGDD86 pKa = 3.59VVPEE90 pKa = 4.2SKK92 pKa = 10.64VAGSPEE98 pKa = 4.09VNLLAPNSQCRR109 pKa = 11.84ILLKK113 pKa = 10.88NSDD116 pKa = 4.23GDD118 pKa = 3.89WVLTGCAVRR127 pKa = 11.84VDD129 pKa = 4.3LPIGPFLVFPQHH141 pKa = 6.02LCAGEE146 pKa = 3.91LAAMSSRR153 pKa = 11.84DD154 pKa = 3.49EE155 pKa = 4.42VIPLSLDD162 pKa = 3.1GLEE165 pKa = 5.16PIAADD170 pKa = 5.11LSMLPCPSKK179 pKa = 10.37LTSTGLASPTIGVVRR194 pKa = 11.84PGQRR198 pKa = 11.84CVVSVVGVDD207 pKa = 3.66GLGTTGPLQTSPVFGRR223 pKa = 11.84LIYY226 pKa = 10.5GATTNTGYY234 pKa = 10.71SGAAYY239 pKa = 10.04ASARR243 pKa = 11.84TVYY246 pKa = 10.17GVHH249 pKa = 6.18TNGGKK254 pKa = 10.53LNGGFALSYY263 pKa = 9.72VYY265 pKa = 11.68AMMKK269 pKa = 9.26VLKK272 pKa = 10.03GVRR275 pKa = 11.84DD276 pKa = 3.64EE277 pKa = 5.43SSEE280 pKa = 3.96NWIHH284 pKa = 7.15SVMADD289 pKa = 2.74KK290 pKa = 10.18STRR293 pKa = 11.84FLEE296 pKa = 3.89MDD298 pKa = 3.68YY299 pKa = 11.35EE300 pKa = 4.22RR301 pKa = 11.84VADD304 pKa = 4.3SIVMRR309 pKa = 11.84GDD311 pKa = 3.11DD312 pKa = 3.96GFYY315 pKa = 10.53HH316 pKa = 7.51AISGPAAEE324 pKa = 4.67GLVKK328 pKa = 10.76KK329 pKa = 10.38LDD331 pKa = 3.48KK332 pKa = 10.84YY333 pKa = 11.12KK334 pKa = 10.77YY335 pKa = 9.2DD336 pKa = 3.23AGMVDD341 pKa = 4.69FRR343 pKa = 11.84SKK345 pKa = 9.61TGHH348 pKa = 5.12TWSDD352 pKa = 2.68IDD354 pKa = 3.9PNHH357 pKa = 6.95WEE359 pKa = 4.08SANPEE364 pKa = 4.16TAQVLASADD373 pKa = 3.59TVPQGFHH380 pKa = 7.29LDD382 pKa = 3.48PSSRR386 pKa = 11.84EE387 pKa = 3.67SPYY390 pKa = 10.25IEE392 pKa = 4.4EE393 pKa = 4.29LASQIRR399 pKa = 11.84MFEE402 pKa = 4.17DD403 pKa = 3.3RR404 pKa = 11.84LKK406 pKa = 11.23SSLVDD411 pKa = 3.29TALKK415 pKa = 9.24KK416 pKa = 10.66QIMDD420 pKa = 4.21TFQQTLEE427 pKa = 4.13PSRR430 pKa = 11.84SLTLSLCRR438 pKa = 11.84PEE440 pKa = 4.27LQHH443 pKa = 7.16RR444 pKa = 11.84LAKK447 pKa = 10.81VMFGLHH453 pKa = 5.64
Molecular weight: 49.33 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1420 |
382 |
585 |
473.3 |
52.89 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.535 ± 0.873 | 1.831 ± 0.21 |
5.282 ± 0.103 | 6.056 ± 0.405 |
4.155 ± 0.559 | 6.549 ± 0.88 |
2.887 ± 0.41 | 3.944 ± 0.518 |
4.718 ± 0.612 | 9.155 ± 0.787 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.465 ± 0.559 | 2.676 ± 0.143 |
5.563 ± 0.877 | 2.958 ± 0.232 |
6.338 ± 0.544 | 9.225 ± 1.416 |
5.845 ± 1.069 | 7.465 ± 0.955 |
1.831 ± 0.601 | 3.521 ± 0.262 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
