
Limnohabitans sp. JirII-31
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Burkholderiales; Comamonadaceae; Limnohabitans; unclassified Limnohabitans
Average proteome isoelectric point is 6.78
Get precalculated fractions of proteins
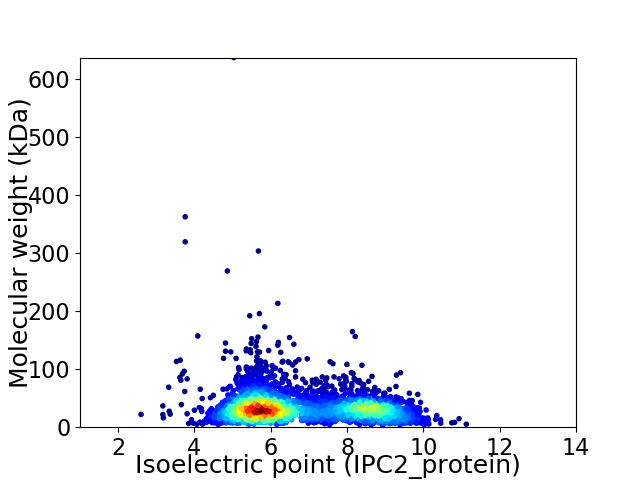
Virtual 2D-PAGE plot for 3283 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
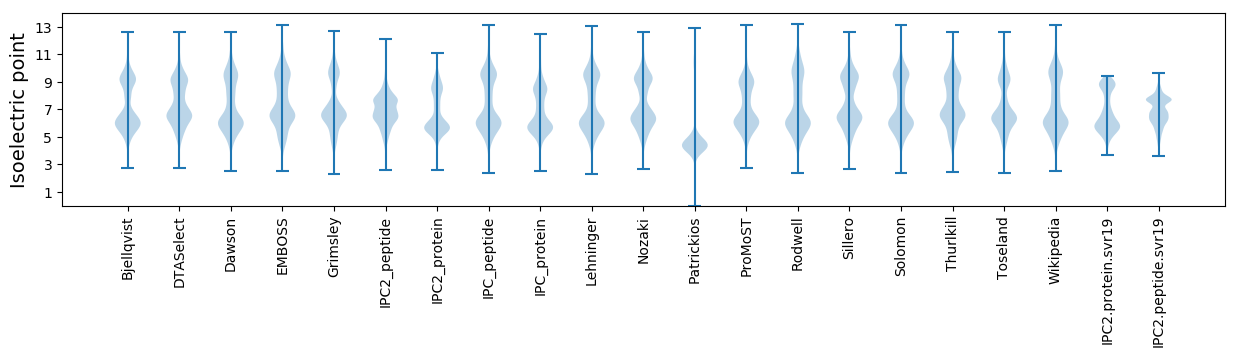
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2M6VIW6|A0A2M6VIW6_9BURK Iron ABC transporter permease OS=Limnohabitans sp. JirII-31 OX=1977908 GN=B9Z41_07990 PE=3 SV=1
MM1 pKa = 7.65SDD3 pKa = 3.31GNGGHH8 pKa = 6.35ATALATINLAAVNDD22 pKa = 3.94APVTVGEE29 pKa = 4.54LSSTDD34 pKa = 3.24EE35 pKa = 4.29DD36 pKa = 3.68VGLTFTQAQLLNNDD50 pKa = 3.87SDD52 pKa = 3.88VDD54 pKa = 3.96TATDD58 pKa = 4.01GQVLTITAVSGATNGTVSMLANGDD82 pKa = 2.88IRR84 pKa = 11.84FIPNANYY91 pKa = 10.23HH92 pKa = 5.27GTASFTYY99 pKa = 9.31TVSDD103 pKa = 4.1GNGGTTDD110 pKa = 3.29ASVSITVNAVNDD122 pKa = 3.79VPVVTGEE129 pKa = 4.32TVSTNEE135 pKa = 4.34DD136 pKa = 3.17TTLWFIPSDD145 pKa = 3.84LLTNDD150 pKa = 3.08TDD152 pKa = 3.96IDD154 pKa = 4.11VATDD158 pKa = 3.79GQVLSITAVSNATHH172 pKa = 5.72GTVGFVTQADD182 pKa = 4.0GSRR185 pKa = 11.84RR186 pKa = 11.84IAFTPDD192 pKa = 2.51ANYY195 pKa = 10.51FGVASFQYY203 pKa = 9.51TVSDD207 pKa = 4.06GAGGTSTGTVSINVAQINDD226 pKa = 3.58VPVASDD232 pKa = 3.82DD233 pKa = 4.03SLSGSAEE240 pKa = 3.96DD241 pKa = 3.76NALHH245 pKa = 7.12ISFAKK250 pKa = 10.53LLSNDD255 pKa = 3.08TDD257 pKa = 3.94ADD259 pKa = 3.81STNALLGGTNDD270 pKa = 3.45VLTVSAVGNATHH282 pKa = 7.14GSVALVNGEE291 pKa = 4.15IVFTPDD297 pKa = 2.75ANYY300 pKa = 10.4HH301 pKa = 5.28GVASFVYY308 pKa = 8.87QVRR311 pKa = 11.84DD312 pKa = 3.23QSGALSQATAAFTITAVNDD331 pKa = 3.48APVATGEE338 pKa = 4.5TISSSEE344 pKa = 4.61DD345 pKa = 3.2SNLLLNQSDD354 pKa = 4.25LLKK357 pKa = 10.73NDD359 pKa = 3.69SDD361 pKa = 3.66VDD363 pKa = 3.67AATDD367 pKa = 4.04GQVLTIKK374 pKa = 10.4AVSNARR380 pKa = 11.84HH381 pKa = 5.79GSVTLNADD389 pKa = 3.3GTILFVPDD397 pKa = 3.26ANYY400 pKa = 11.03YY401 pKa = 8.07GTASFDD407 pKa = 3.58YY408 pKa = 10.64TVDD411 pKa = 3.84DD412 pKa = 4.87GNGGAASATATITLLSVNDD431 pKa = 3.86APVATGEE438 pKa = 4.36TVQDD442 pKa = 4.06VEE444 pKa = 4.44DD445 pKa = 3.66QQLTILSSALLQNDD459 pKa = 3.42TDD461 pKa = 3.63VDD463 pKa = 3.72NTQASLTISRR473 pKa = 11.84VLSGAGGTAYY483 pKa = 10.81LNASGHH489 pKa = 5.67VVFTPNTNYY498 pKa = 10.7NGNATFTYY506 pKa = 8.86WVKK509 pKa = 11.14DD510 pKa = 3.46PDD512 pKa = 4.06GLEE515 pKa = 4.38SNAVTATVVVSAVNDD530 pKa = 3.44APTAQGEE537 pKa = 4.64IVTGASEE544 pKa = 4.55DD545 pKa = 3.45AVFYY549 pKa = 10.69INKK552 pKa = 8.37STLLANDD559 pKa = 3.91SDD561 pKa = 4.31IDD563 pKa = 4.03DD564 pKa = 4.43ASSALSLSWVGNASGGSVSLDD585 pKa = 3.13GNGNVVFTPNANYY598 pKa = 10.45NGNASFQYY606 pKa = 10.14KK607 pKa = 9.64VRR609 pKa = 11.84DD610 pKa = 3.39AAGAEE615 pKa = 4.38SAVVQAVIPVTAVNDD630 pKa = 3.67APVAVDD636 pKa = 5.57DD637 pKa = 6.03KK638 pKa = 11.56FDD640 pKa = 3.68TYY642 pKa = 11.64KK643 pKa = 9.69NTPVTVAFNQLTSNDD658 pKa = 3.52SDD660 pKa = 3.78VDD662 pKa = 3.84GDD664 pKa = 4.29SLTVSAVRR672 pKa = 11.84DD673 pKa = 3.69HH674 pKa = 7.15ANGHH678 pKa = 5.74ASIVNGQVQFQATTGFTGTASFDD701 pKa = 3.62YY702 pKa = 10.96LSDD705 pKa = 4.67DD706 pKa = 3.56GHH708 pKa = 7.17GGQTWATAYY717 pKa = 10.87VNVKK721 pKa = 9.7VPPNQYY727 pKa = 8.55PTMTLKK733 pKa = 10.69YY734 pKa = 10.56FPVCDD739 pKa = 3.58GKK741 pKa = 11.16YY742 pKa = 10.44GYY744 pKa = 10.06DD745 pKa = 3.14ILGSTILTNGIQTFSLHH762 pKa = 7.48DD763 pKa = 4.9DD764 pKa = 3.98GSTSAISMRR773 pKa = 11.84FVSATFKK780 pKa = 10.83PPLVLTLL787 pKa = 3.95
MM1 pKa = 7.65SDD3 pKa = 3.31GNGGHH8 pKa = 6.35ATALATINLAAVNDD22 pKa = 3.94APVTVGEE29 pKa = 4.54LSSTDD34 pKa = 3.24EE35 pKa = 4.29DD36 pKa = 3.68VGLTFTQAQLLNNDD50 pKa = 3.87SDD52 pKa = 3.88VDD54 pKa = 3.96TATDD58 pKa = 4.01GQVLTITAVSGATNGTVSMLANGDD82 pKa = 2.88IRR84 pKa = 11.84FIPNANYY91 pKa = 10.23HH92 pKa = 5.27GTASFTYY99 pKa = 9.31TVSDD103 pKa = 4.1GNGGTTDD110 pKa = 3.29ASVSITVNAVNDD122 pKa = 3.79VPVVTGEE129 pKa = 4.32TVSTNEE135 pKa = 4.34DD136 pKa = 3.17TTLWFIPSDD145 pKa = 3.84LLTNDD150 pKa = 3.08TDD152 pKa = 3.96IDD154 pKa = 4.11VATDD158 pKa = 3.79GQVLSITAVSNATHH172 pKa = 5.72GTVGFVTQADD182 pKa = 4.0GSRR185 pKa = 11.84RR186 pKa = 11.84IAFTPDD192 pKa = 2.51ANYY195 pKa = 10.51FGVASFQYY203 pKa = 9.51TVSDD207 pKa = 4.06GAGGTSTGTVSINVAQINDD226 pKa = 3.58VPVASDD232 pKa = 3.82DD233 pKa = 4.03SLSGSAEE240 pKa = 3.96DD241 pKa = 3.76NALHH245 pKa = 7.12ISFAKK250 pKa = 10.53LLSNDD255 pKa = 3.08TDD257 pKa = 3.94ADD259 pKa = 3.81STNALLGGTNDD270 pKa = 3.45VLTVSAVGNATHH282 pKa = 7.14GSVALVNGEE291 pKa = 4.15IVFTPDD297 pKa = 2.75ANYY300 pKa = 10.4HH301 pKa = 5.28GVASFVYY308 pKa = 8.87QVRR311 pKa = 11.84DD312 pKa = 3.23QSGALSQATAAFTITAVNDD331 pKa = 3.48APVATGEE338 pKa = 4.5TISSSEE344 pKa = 4.61DD345 pKa = 3.2SNLLLNQSDD354 pKa = 4.25LLKK357 pKa = 10.73NDD359 pKa = 3.69SDD361 pKa = 3.66VDD363 pKa = 3.67AATDD367 pKa = 4.04GQVLTIKK374 pKa = 10.4AVSNARR380 pKa = 11.84HH381 pKa = 5.79GSVTLNADD389 pKa = 3.3GTILFVPDD397 pKa = 3.26ANYY400 pKa = 11.03YY401 pKa = 8.07GTASFDD407 pKa = 3.58YY408 pKa = 10.64TVDD411 pKa = 3.84DD412 pKa = 4.87GNGGAASATATITLLSVNDD431 pKa = 3.86APVATGEE438 pKa = 4.36TVQDD442 pKa = 4.06VEE444 pKa = 4.44DD445 pKa = 3.66QQLTILSSALLQNDD459 pKa = 3.42TDD461 pKa = 3.63VDD463 pKa = 3.72NTQASLTISRR473 pKa = 11.84VLSGAGGTAYY483 pKa = 10.81LNASGHH489 pKa = 5.67VVFTPNTNYY498 pKa = 10.7NGNATFTYY506 pKa = 8.86WVKK509 pKa = 11.14DD510 pKa = 3.46PDD512 pKa = 4.06GLEE515 pKa = 4.38SNAVTATVVVSAVNDD530 pKa = 3.44APTAQGEE537 pKa = 4.64IVTGASEE544 pKa = 4.55DD545 pKa = 3.45AVFYY549 pKa = 10.69INKK552 pKa = 8.37STLLANDD559 pKa = 3.91SDD561 pKa = 4.31IDD563 pKa = 4.03DD564 pKa = 4.43ASSALSLSWVGNASGGSVSLDD585 pKa = 3.13GNGNVVFTPNANYY598 pKa = 10.45NGNASFQYY606 pKa = 10.14KK607 pKa = 9.64VRR609 pKa = 11.84DD610 pKa = 3.39AAGAEE615 pKa = 4.38SAVVQAVIPVTAVNDD630 pKa = 3.67APVAVDD636 pKa = 5.57DD637 pKa = 6.03KK638 pKa = 11.56FDD640 pKa = 3.68TYY642 pKa = 11.64KK643 pKa = 9.69NTPVTVAFNQLTSNDD658 pKa = 3.52SDD660 pKa = 3.78VDD662 pKa = 3.84GDD664 pKa = 4.29SLTVSAVRR672 pKa = 11.84DD673 pKa = 3.69HH674 pKa = 7.15ANGHH678 pKa = 5.74ASIVNGQVQFQATTGFTGTASFDD701 pKa = 3.62YY702 pKa = 10.96LSDD705 pKa = 4.67DD706 pKa = 3.56GHH708 pKa = 7.17GGQTWATAYY717 pKa = 10.87VNVKK721 pKa = 9.7VPPNQYY727 pKa = 8.55PTMTLKK733 pKa = 10.69YY734 pKa = 10.56FPVCDD739 pKa = 3.58GKK741 pKa = 11.16YY742 pKa = 10.44GYY744 pKa = 10.06DD745 pKa = 3.14ILGSTILTNGIQTFSLHH762 pKa = 7.48DD763 pKa = 4.9DD764 pKa = 3.98GSTSAISMRR773 pKa = 11.84FVSATFKK780 pKa = 10.83PPLVLTLL787 pKa = 3.95
Molecular weight: 80.85 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2M6VJ29|A0A2M6VJ29_9BURK zf-CHCC domain-containing protein OS=Limnohabitans sp. JirII-31 OX=1977908 GN=B9Z41_08605 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.25QPSKK9 pKa = 7.79TRR11 pKa = 11.84RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.79GFLVRR21 pKa = 11.84MKK23 pKa = 9.7TRR25 pKa = 11.84GGRR28 pKa = 11.84AVINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.58GRR39 pKa = 11.84KK40 pKa = 8.75RR41 pKa = 11.84LAVV44 pKa = 3.41
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.25QPSKK9 pKa = 7.79TRR11 pKa = 11.84RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.79GFLVRR21 pKa = 11.84MKK23 pKa = 9.7TRR25 pKa = 11.84GGRR28 pKa = 11.84AVINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.58GRR39 pKa = 11.84KK40 pKa = 8.75RR41 pKa = 11.84LAVV44 pKa = 3.41
Molecular weight: 5.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1066846 |
33 |
6459 |
325.0 |
35.39 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.608 ± 0.051 | 0.977 ± 0.016 |
5.235 ± 0.039 | 4.949 ± 0.046 |
3.542 ± 0.033 | 7.886 ± 0.055 |
2.596 ± 0.03 | 4.454 ± 0.031 |
3.849 ± 0.047 | 10.565 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.71 ± 0.026 | 3.092 ± 0.052 |
4.963 ± 0.036 | 4.616 ± 0.037 |
5.737 ± 0.043 | 5.828 ± 0.057 |
5.636 ± 0.072 | 7.956 ± 0.036 |
1.492 ± 0.022 | 2.31 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
