
Paramuribaculum intestinale
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Bacteroidia; Bacteroidales; Muribaculaceae; Paramuribaculum
Average proteome isoelectric point is 6.37
Get precalculated fractions of proteins
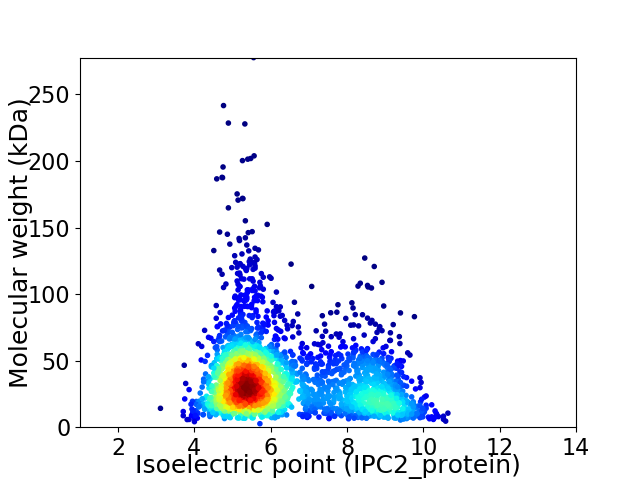
Virtual 2D-PAGE plot for 2513 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2V1IS55|A0A2V1IS55_9BACT Cell filamentation protein Fic OS=Paramuribaculum intestinale OX=2094151 GN=C5O25_12515 PE=4 SV=1
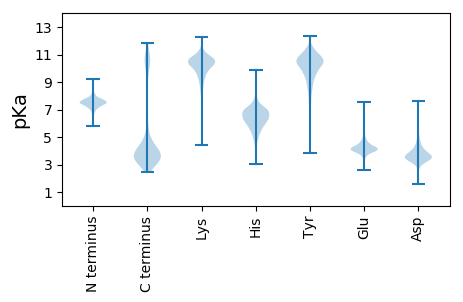
MM1 pKa = 7.7KK2 pKa = 10.27AFHH5 pKa = 6.41YY6 pKa = 10.09LAAAAALLFAFTAASCSDD24 pKa = 3.64SNEE27 pKa = 3.86EE28 pKa = 4.5PEE30 pKa = 4.38PEE32 pKa = 4.61VINHH36 pKa = 6.79IYY38 pKa = 10.61FSIPDD43 pKa = 4.04GNSSITDD50 pKa = 3.49NTDD53 pKa = 2.79EE54 pKa = 4.43TVTVAISLSSPLATATSFTVEE75 pKa = 4.17LTGSDD80 pKa = 3.59ASLLTIGNNPVSIAAGSTTGTFTVASANAGTITEE114 pKa = 4.32STSFGFTIRR123 pKa = 11.84DD124 pKa = 3.41LDD126 pKa = 3.78PAKK129 pKa = 10.57FDD131 pKa = 3.24IAQNATVTLLPAANEE146 pKa = 4.2STLTPEE152 pKa = 3.83EE153 pKa = 3.97QALVDD158 pKa = 3.44QWRR161 pKa = 11.84TAYY164 pKa = 10.42GIDD167 pKa = 3.42LTPWIGTVALQGTIEE182 pKa = 4.43FPGSGNRR189 pKa = 11.84APFISPQTIQLSGSTVFAISKK210 pKa = 10.17DD211 pKa = 3.63ADD213 pKa = 3.6DD214 pKa = 4.16TTPVLDD220 pKa = 3.54MTEE223 pKa = 4.35NPMGMTEE230 pKa = 4.1YY231 pKa = 10.59LYY233 pKa = 9.55KK234 pKa = 10.16TLRR237 pKa = 11.84QLTVDD242 pKa = 3.56DD243 pKa = 4.41NEE245 pKa = 4.22YY246 pKa = 10.66FALEE250 pKa = 4.27DD251 pKa = 4.41DD252 pKa = 4.47GSGLEE257 pKa = 4.8LMEE260 pKa = 5.62LINWNAKK267 pKa = 9.02SEE269 pKa = 4.13EE270 pKa = 4.42SFTVSLPGLKK280 pKa = 8.55ITDD283 pKa = 3.62IQDD286 pKa = 3.08GKK288 pKa = 10.32ATVEE292 pKa = 4.65FVAEE296 pKa = 4.09GDD298 pKa = 3.97DD299 pKa = 4.46YY300 pKa = 11.57ILNAQGEE307 pKa = 4.82PIYY310 pKa = 11.04SPNTDD315 pKa = 2.98EE316 pKa = 4.3YY317 pKa = 10.0LTYY320 pKa = 10.05YY321 pKa = 9.52YY322 pKa = 10.05HH323 pKa = 7.26SSWIPFSYY331 pKa = 10.53QFTAWNRR338 pKa = 11.84QLALIEE344 pKa = 4.49NNDD347 pKa = 4.36PIAMEE352 pKa = 4.01MQSYY356 pKa = 9.97GISAAPATHH365 pKa = 7.0LGLSDD370 pKa = 3.59VLEE373 pKa = 4.91DD374 pKa = 3.08SWEE377 pKa = 3.86IDD379 pKa = 3.23EE380 pKa = 6.36DD381 pKa = 4.11EE382 pKa = 5.48DD383 pKa = 4.7GVSNLYY389 pKa = 9.27VAPHH393 pKa = 6.65AEE395 pKa = 3.66IDD397 pKa = 4.09FNAGTMTFIFPFDD410 pKa = 3.7TEE412 pKa = 4.07DD413 pKa = 3.14QYY415 pKa = 11.96GYY417 pKa = 11.25SRR419 pKa = 11.84ITVTYY424 pKa = 8.58TLQKK428 pKa = 10.84
MM1 pKa = 7.7KK2 pKa = 10.27AFHH5 pKa = 6.41YY6 pKa = 10.09LAAAAALLFAFTAASCSDD24 pKa = 3.64SNEE27 pKa = 3.86EE28 pKa = 4.5PEE30 pKa = 4.38PEE32 pKa = 4.61VINHH36 pKa = 6.79IYY38 pKa = 10.61FSIPDD43 pKa = 4.04GNSSITDD50 pKa = 3.49NTDD53 pKa = 2.79EE54 pKa = 4.43TVTVAISLSSPLATATSFTVEE75 pKa = 4.17LTGSDD80 pKa = 3.59ASLLTIGNNPVSIAAGSTTGTFTVASANAGTITEE114 pKa = 4.32STSFGFTIRR123 pKa = 11.84DD124 pKa = 3.41LDD126 pKa = 3.78PAKK129 pKa = 10.57FDD131 pKa = 3.24IAQNATVTLLPAANEE146 pKa = 4.2STLTPEE152 pKa = 3.83EE153 pKa = 3.97QALVDD158 pKa = 3.44QWRR161 pKa = 11.84TAYY164 pKa = 10.42GIDD167 pKa = 3.42LTPWIGTVALQGTIEE182 pKa = 4.43FPGSGNRR189 pKa = 11.84APFISPQTIQLSGSTVFAISKK210 pKa = 10.17DD211 pKa = 3.63ADD213 pKa = 3.6DD214 pKa = 4.16TTPVLDD220 pKa = 3.54MTEE223 pKa = 4.35NPMGMTEE230 pKa = 4.1YY231 pKa = 10.59LYY233 pKa = 9.55KK234 pKa = 10.16TLRR237 pKa = 11.84QLTVDD242 pKa = 3.56DD243 pKa = 4.41NEE245 pKa = 4.22YY246 pKa = 10.66FALEE250 pKa = 4.27DD251 pKa = 4.41DD252 pKa = 4.47GSGLEE257 pKa = 4.8LMEE260 pKa = 5.62LINWNAKK267 pKa = 9.02SEE269 pKa = 4.13EE270 pKa = 4.42SFTVSLPGLKK280 pKa = 8.55ITDD283 pKa = 3.62IQDD286 pKa = 3.08GKK288 pKa = 10.32ATVEE292 pKa = 4.65FVAEE296 pKa = 4.09GDD298 pKa = 3.97DD299 pKa = 4.46YY300 pKa = 11.57ILNAQGEE307 pKa = 4.82PIYY310 pKa = 11.04SPNTDD315 pKa = 2.98EE316 pKa = 4.3YY317 pKa = 10.0LTYY320 pKa = 10.05YY321 pKa = 9.52YY322 pKa = 10.05HH323 pKa = 7.26SSWIPFSYY331 pKa = 10.53QFTAWNRR338 pKa = 11.84QLALIEE344 pKa = 4.49NNDD347 pKa = 4.36PIAMEE352 pKa = 4.01MQSYY356 pKa = 9.97GISAAPATHH365 pKa = 7.0LGLSDD370 pKa = 3.59VLEE373 pKa = 4.91DD374 pKa = 3.08SWEE377 pKa = 3.86IDD379 pKa = 3.23EE380 pKa = 6.36DD381 pKa = 4.11EE382 pKa = 5.48DD383 pKa = 4.7GVSNLYY389 pKa = 9.27VAPHH393 pKa = 6.65AEE395 pKa = 3.66IDD397 pKa = 4.09FNAGTMTFIFPFDD410 pKa = 3.7TEE412 pKa = 4.07DD413 pKa = 3.14QYY415 pKa = 11.96GYY417 pKa = 11.25SRR419 pKa = 11.84ITVTYY424 pKa = 8.58TLQKK428 pKa = 10.84
Molecular weight: 46.56 kDa
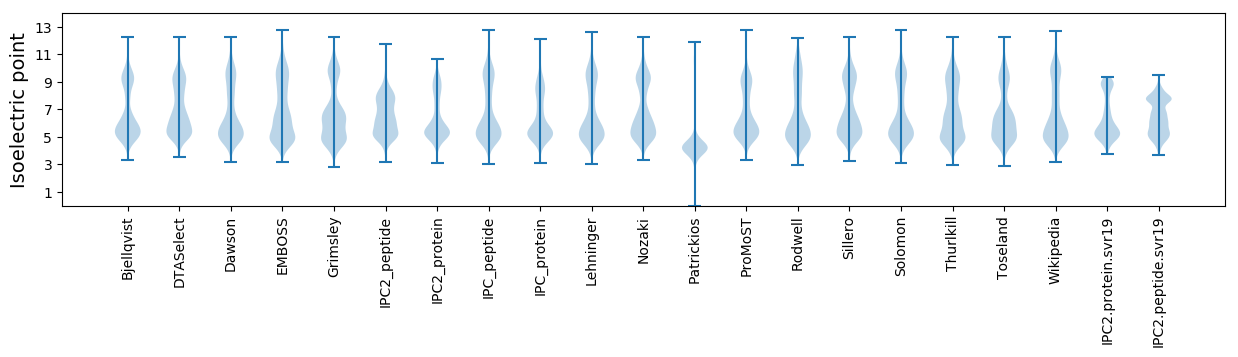
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2V1J1V4|A0A2V1J1V4_9BACT Uncharacterized protein OS=Paramuribaculum intestinale OX=2094151 GN=C5O25_03945 PE=4 SV=1
MM1 pKa = 7.57SYY3 pKa = 11.29VDD5 pKa = 4.45IILIVLALGAAVIGYY20 pKa = 9.27RR21 pKa = 11.84RR22 pKa = 11.84GAIRR26 pKa = 11.84QAASIGGILVALFAVRR42 pKa = 11.84MAGDD46 pKa = 3.43KK47 pKa = 10.93ASAAVAGMIGQSPADD62 pKa = 3.59YY63 pKa = 10.06ASRR66 pKa = 11.84IIGCGLLFVAVWGGCWLLGRR86 pKa = 11.84MLRR89 pKa = 11.84LSVRR93 pKa = 11.84AVMLGAVDD101 pKa = 4.27SVAGSVFLIFKK112 pKa = 9.11WGLVVSLLLNLWRR125 pKa = 11.84VVSPDD130 pKa = 2.73SGIFVSSRR138 pKa = 11.84LMGGRR143 pKa = 11.84IFAAVMDD150 pKa = 4.27MAPAVMGFLRR160 pKa = 11.84DD161 pKa = 3.57SFMGASS167 pKa = 3.38
MM1 pKa = 7.57SYY3 pKa = 11.29VDD5 pKa = 4.45IILIVLALGAAVIGYY20 pKa = 9.27RR21 pKa = 11.84RR22 pKa = 11.84GAIRR26 pKa = 11.84QAASIGGILVALFAVRR42 pKa = 11.84MAGDD46 pKa = 3.43KK47 pKa = 10.93ASAAVAGMIGQSPADD62 pKa = 3.59YY63 pKa = 10.06ASRR66 pKa = 11.84IIGCGLLFVAVWGGCWLLGRR86 pKa = 11.84MLRR89 pKa = 11.84LSVRR93 pKa = 11.84AVMLGAVDD101 pKa = 4.27SVAGSVFLIFKK112 pKa = 9.11WGLVVSLLLNLWRR125 pKa = 11.84VVSPDD130 pKa = 2.73SGIFVSSRR138 pKa = 11.84LMGGRR143 pKa = 11.84IFAAVMDD150 pKa = 4.27MAPAVMGFLRR160 pKa = 11.84DD161 pKa = 3.57SFMGASS167 pKa = 3.38
Molecular weight: 17.47 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
863129 |
25 |
2477 |
343.5 |
38.3 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.783 ± 0.057 | 1.229 ± 0.02 |
6.298 ± 0.031 | 5.964 ± 0.043 |
3.997 ± 0.032 | 7.075 ± 0.041 |
1.99 ± 0.022 | 6.584 ± 0.038 |
4.946 ± 0.049 | 8.772 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.929 ± 0.021 | 4.189 ± 0.042 |
4.185 ± 0.031 | 3.032 ± 0.028 |
6.197 ± 0.041 | 6.397 ± 0.038 |
5.824 ± 0.043 | 6.686 ± 0.042 |
1.189 ± 0.022 | 3.733 ± 0.032 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
