
Sweet potato symptomless virus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Geminiviridae; Mastrevirus
Average proteome isoelectric point is 8.06
Get precalculated fractions of proteins
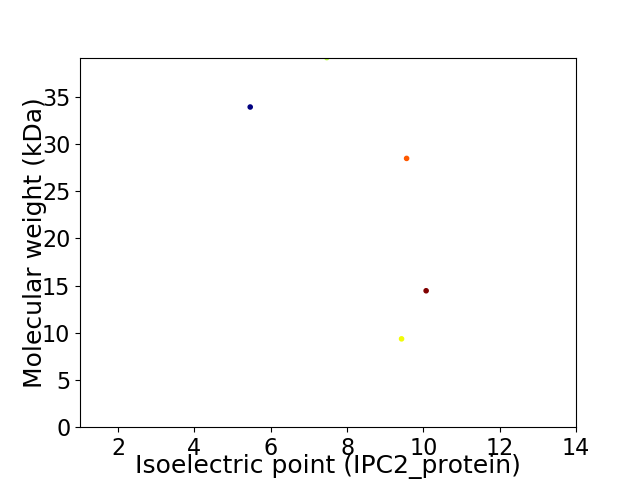
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
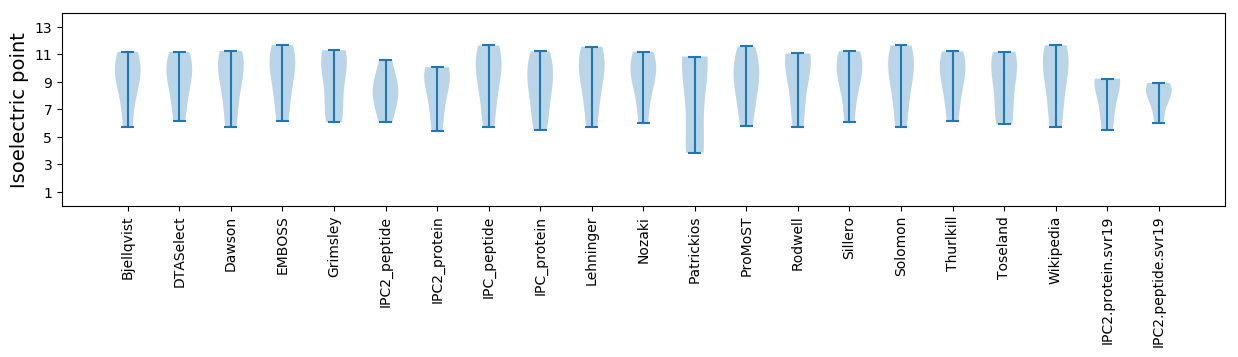
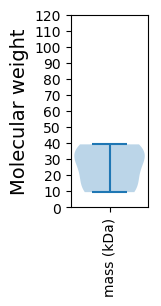
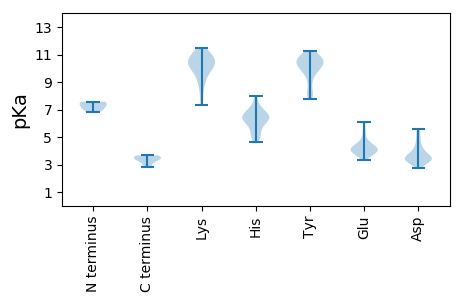
Protein with the lowest isoelectric point:
>tr|A0A2Z2GUW7|A0A2Z2GUW7_9GEMI Isoform of A0A2Z2GPB7 Replication-associated protein OS=Sweet potato symptomless virus 1 OX=603333 GN=C1 PE=3 SV=1
MM1 pKa = 7.57PRR3 pKa = 11.84QPRR6 pKa = 11.84ARR8 pKa = 11.84SVFSFSSKK16 pKa = 10.93YY17 pKa = 10.87GFLTYY22 pKa = 9.81SQCDD26 pKa = 3.51LTAEE30 pKa = 5.44IIQQMLITLLTPFNLLFLAVAPEE53 pKa = 3.8HH54 pKa = 6.4HH55 pKa = 6.77QDD57 pKa = 3.29GSLHH61 pKa = 5.04FHH63 pKa = 6.59VLFQCARR70 pKa = 11.84RR71 pKa = 11.84IITRR75 pKa = 11.84RR76 pKa = 11.84VDD78 pKa = 5.13FFDD81 pKa = 4.53LNNYY85 pKa = 9.02HH86 pKa = 7.26PNIQPARR93 pKa = 11.84DD94 pKa = 3.35SAAVLEE100 pKa = 4.86YY101 pKa = 10.06ISKK104 pKa = 8.8EE105 pKa = 3.66QPPIVWGEE113 pKa = 3.86FQNHH117 pKa = 5.26KK118 pKa = 10.0VSPSRR123 pKa = 11.84RR124 pKa = 11.84DD125 pKa = 3.41DD126 pKa = 2.92RR127 pKa = 11.84WRR129 pKa = 11.84DD130 pKa = 3.42IIFTSTSKK138 pKa = 11.14AEE140 pKa = 3.78YY141 pKa = 10.06LNRR144 pKa = 11.84VKK146 pKa = 10.73QEE148 pKa = 3.95FPADD152 pKa = 3.41YY153 pKa = 8.2ATRR156 pKa = 11.84LQQLEE161 pKa = 4.41YY162 pKa = 9.88FAEE165 pKa = 4.14RR166 pKa = 11.84LFPSTAPPYY175 pKa = 10.46VSPFDD180 pKa = 4.21PLTLQCHH187 pKa = 6.24EE188 pKa = 6.13DD189 pKa = 3.21IQDD192 pKa = 3.9WINRR196 pKa = 11.84EE197 pKa = 3.9LYY199 pKa = 10.19LVHH202 pKa = 7.99LEE204 pKa = 4.53DD205 pKa = 5.4YY206 pKa = 10.82ILIHH210 pKa = 6.11NCSYY214 pKa = 10.1DD215 pKa = 3.32TARR218 pKa = 11.84QDD220 pKa = 5.57LEE222 pKa = 4.21WLDD225 pKa = 4.08DD226 pKa = 3.94LARR229 pKa = 11.84NPVKK233 pKa = 10.44EE234 pKa = 4.13PSEE237 pKa = 4.42DD238 pKa = 3.81PGPSTYY244 pKa = 10.39VVPPAQQRR252 pKa = 11.84QPGQDD257 pKa = 3.06NSGGTTTGTVGLTSPRR273 pKa = 11.84TTVMPHH279 pKa = 4.65TTSPTTSHH287 pKa = 6.57SNSSQAGSNN296 pKa = 3.38
MM1 pKa = 7.57PRR3 pKa = 11.84QPRR6 pKa = 11.84ARR8 pKa = 11.84SVFSFSSKK16 pKa = 10.93YY17 pKa = 10.87GFLTYY22 pKa = 9.81SQCDD26 pKa = 3.51LTAEE30 pKa = 5.44IIQQMLITLLTPFNLLFLAVAPEE53 pKa = 3.8HH54 pKa = 6.4HH55 pKa = 6.77QDD57 pKa = 3.29GSLHH61 pKa = 5.04FHH63 pKa = 6.59VLFQCARR70 pKa = 11.84RR71 pKa = 11.84IITRR75 pKa = 11.84RR76 pKa = 11.84VDD78 pKa = 5.13FFDD81 pKa = 4.53LNNYY85 pKa = 9.02HH86 pKa = 7.26PNIQPARR93 pKa = 11.84DD94 pKa = 3.35SAAVLEE100 pKa = 4.86YY101 pKa = 10.06ISKK104 pKa = 8.8EE105 pKa = 3.66QPPIVWGEE113 pKa = 3.86FQNHH117 pKa = 5.26KK118 pKa = 10.0VSPSRR123 pKa = 11.84RR124 pKa = 11.84DD125 pKa = 3.41DD126 pKa = 2.92RR127 pKa = 11.84WRR129 pKa = 11.84DD130 pKa = 3.42IIFTSTSKK138 pKa = 11.14AEE140 pKa = 3.78YY141 pKa = 10.06LNRR144 pKa = 11.84VKK146 pKa = 10.73QEE148 pKa = 3.95FPADD152 pKa = 3.41YY153 pKa = 8.2ATRR156 pKa = 11.84LQQLEE161 pKa = 4.41YY162 pKa = 9.88FAEE165 pKa = 4.14RR166 pKa = 11.84LFPSTAPPYY175 pKa = 10.46VSPFDD180 pKa = 4.21PLTLQCHH187 pKa = 6.24EE188 pKa = 6.13DD189 pKa = 3.21IQDD192 pKa = 3.9WINRR196 pKa = 11.84EE197 pKa = 3.9LYY199 pKa = 10.19LVHH202 pKa = 7.99LEE204 pKa = 4.53DD205 pKa = 5.4YY206 pKa = 10.82ILIHH210 pKa = 6.11NCSYY214 pKa = 10.1DD215 pKa = 3.32TARR218 pKa = 11.84QDD220 pKa = 5.57LEE222 pKa = 4.21WLDD225 pKa = 4.08DD226 pKa = 3.94LARR229 pKa = 11.84NPVKK233 pKa = 10.44EE234 pKa = 4.13PSEE237 pKa = 4.42DD238 pKa = 3.81PGPSTYY244 pKa = 10.39VVPPAQQRR252 pKa = 11.84QPGQDD257 pKa = 3.06NSGGTTTGTVGLTSPRR273 pKa = 11.84TTVMPHH279 pKa = 4.65TTSPTTSHH287 pKa = 6.57SNSSQAGSNN296 pKa = 3.38
Molecular weight: 33.91 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2U8ZUC4|A0A2U8ZUC4_9GEMI Capsid protein OS=Sweet potato symptomless virus 1 OX=603333 GN=V2 PE=3 SV=1
MM1 pKa = 7.13VFSPTLNVISQLRR14 pKa = 11.84SFSKK18 pKa = 10.63CLSLSSHH25 pKa = 6.46LLIFSFSLLLLNIIRR40 pKa = 11.84MVPSIFTFSFNVLVGLSLVGSISLILIITIQTYY73 pKa = 10.64NLLVTLQQSLNTYY86 pKa = 8.02QRR88 pKa = 11.84NNLLSSGGNSRR99 pKa = 11.84IIKK102 pKa = 9.36YY103 pKa = 9.77RR104 pKa = 11.84LHH106 pKa = 6.92EE107 pKa = 4.45GMTDD111 pKa = 3.71GEE113 pKa = 4.23ISSLRR118 pKa = 11.84RR119 pKa = 11.84RR120 pKa = 11.84LRR122 pKa = 11.84RR123 pKa = 11.84STLTEE128 pKa = 3.67
MM1 pKa = 7.13VFSPTLNVISQLRR14 pKa = 11.84SFSKK18 pKa = 10.63CLSLSSHH25 pKa = 6.46LLIFSFSLLLLNIIRR40 pKa = 11.84MVPSIFTFSFNVLVGLSLVGSISLILIITIQTYY73 pKa = 10.64NLLVTLQQSLNTYY86 pKa = 8.02QRR88 pKa = 11.84NNLLSSGGNSRR99 pKa = 11.84IIKK102 pKa = 9.36YY103 pKa = 9.77RR104 pKa = 11.84LHH106 pKa = 6.92EE107 pKa = 4.45GMTDD111 pKa = 3.71GEE113 pKa = 4.23ISSLRR118 pKa = 11.84RR119 pKa = 11.84RR120 pKa = 11.84LRR122 pKa = 11.84RR123 pKa = 11.84STLTEE128 pKa = 3.67
Molecular weight: 14.45 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
1097 |
86 |
335 |
219.4 |
25.07 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.105 ± 0.882 | 1.459 ± 0.286 |
5.014 ± 0.837 | 3.464 ± 0.769 |
5.561 ± 0.346 | 5.105 ± 1.051 |
2.37 ± 0.585 | 6.016 ± 0.672 |
4.284 ± 1.094 | 9.116 ± 1.657 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.641 ± 0.346 | 4.284 ± 0.3 |
6.381 ± 1.05 | 5.469 ± 0.475 |
6.837 ± 0.352 | 8.295 ± 1.252 |
7.475 ± 0.582 | 6.29 ± 1.213 |
1.641 ± 0.407 | 4.193 ± 0.334 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
