
Saccharomonospora marina XMU15
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Pseudonocardiales; Pseudonocardiaceae; Saccharomonospora; Saccharomonospora marina
Average proteome isoelectric point is 6.39
Get precalculated fractions of proteins
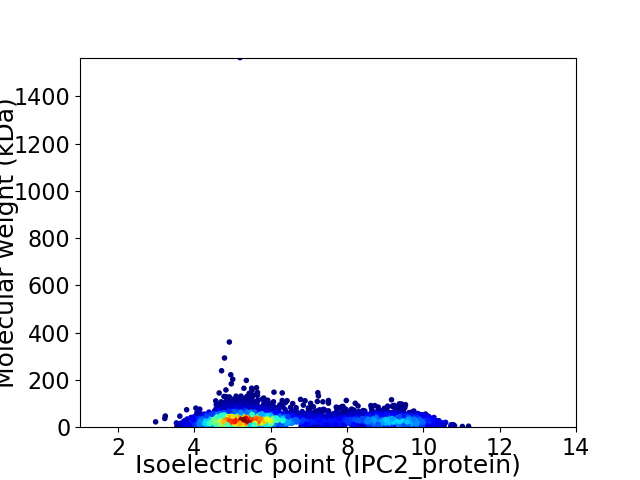
Virtual 2D-PAGE plot for 5567 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|H5X1M8|H5X1M8_9PSEU Enoyl-CoA hydratase/carnithine racemase OS=Saccharomonospora marina XMU15 OX=882083 GN=SacmaDRAFT_2650 PE=3 SV=1
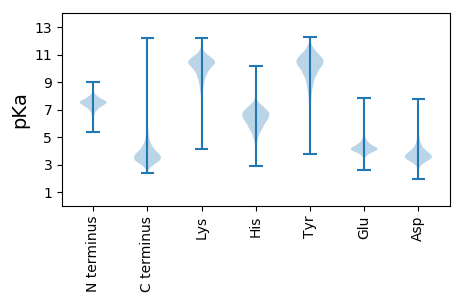
MM1 pKa = 7.45YY2 pKa = 10.59SRR4 pKa = 11.84LPSATPRR11 pKa = 11.84KK12 pKa = 8.49PRR14 pKa = 11.84PIARR18 pKa = 11.84LRR20 pKa = 11.84AAAGRR25 pKa = 11.84FAVAILGLLVATLWLAPASAAPRR48 pKa = 11.84TPEE51 pKa = 3.95PAAGTAQAQVLTWTANDD68 pKa = 4.45SITSYY73 pKa = 11.52ASAPSTATAGAATIVFEE90 pKa = 4.62NSTATGNTTGLPHH103 pKa = 7.24TLTFDD108 pKa = 3.43TSTPGYY114 pKa = 9.95NHH116 pKa = 7.1DD117 pKa = 3.5VDD119 pKa = 4.74VNILASPFDD128 pKa = 4.28ANEE131 pKa = 3.25GRR133 pKa = 11.84YY134 pKa = 8.55EE135 pKa = 4.18VNVNLSPGEE144 pKa = 3.83YY145 pKa = 9.14RR146 pKa = 11.84YY147 pKa = 10.47FCDD150 pKa = 3.89MPGHH154 pKa = 5.67GQMTGVLVVTGEE166 pKa = 4.37GSDD169 pKa = 3.39TTAPEE174 pKa = 3.79VSAEE178 pKa = 4.13VTGQTDD184 pKa = 3.25DD185 pKa = 3.13QGNYY189 pKa = 9.69LGSATVSVTATDD201 pKa = 3.39AGSGVASVEE210 pKa = 3.93YY211 pKa = 10.35EE212 pKa = 3.64IDD214 pKa = 3.43DD215 pKa = 4.21TGFQPYY221 pKa = 8.77TGPVTVDD228 pKa = 2.75RR229 pKa = 11.84VGDD232 pKa = 3.52HH233 pKa = 5.81SVQYY237 pKa = 10.47RR238 pKa = 11.84ATDD241 pKa = 3.44NAGNTSPTGSVQFSVVEE258 pKa = 4.12PDD260 pKa = 4.2PGDD263 pKa = 3.48TTPPEE268 pKa = 3.96VSAEE272 pKa = 4.16VGGEE276 pKa = 3.69TDD278 pKa = 3.1GQGNYY283 pKa = 9.72VGSATVTVAATDD295 pKa = 3.55TGSGVASVEE304 pKa = 4.06YY305 pKa = 9.95EE306 pKa = 3.62LDD308 pKa = 3.49GGGFRR313 pKa = 11.84TYY315 pKa = 10.45SAPVVVDD322 pKa = 3.11TEE324 pKa = 4.52GEE326 pKa = 4.12HH327 pKa = 4.82VVRR330 pKa = 11.84YY331 pKa = 9.11RR332 pKa = 11.84ATDD335 pKa = 3.18NAGNVSQSGSVSFTVVPPDD354 pKa = 4.55PGDD357 pKa = 3.45TTPPQVTAEE366 pKa = 4.0IAGEE370 pKa = 3.8RR371 pKa = 11.84DD372 pKa = 2.8AGGNYY377 pKa = 9.87LGSATVTLAAQDD389 pKa = 4.02SEE391 pKa = 4.87SQVDD395 pKa = 4.03SIEE398 pKa = 4.0YY399 pKa = 10.38AIGDD403 pKa = 4.06GAFQAYY409 pKa = 8.09AAPVTFDD416 pKa = 3.15QPGEE420 pKa = 3.82YY421 pKa = 9.05TLRR424 pKa = 11.84YY425 pKa = 9.21RR426 pKa = 11.84ATDD429 pKa = 3.29TAGNTSQPGSVSFAVVEE446 pKa = 4.34PPPDD450 pKa = 3.61DD451 pKa = 3.41TTAPEE456 pKa = 3.99VSAEE460 pKa = 4.15VGGEE464 pKa = 3.69TDD466 pKa = 3.1GQGNYY471 pKa = 9.72VGSATVTISASDD483 pKa = 3.76ADD485 pKa = 3.92SGIAAVEE492 pKa = 4.06YY493 pKa = 10.82ALDD496 pKa = 3.95DD497 pKa = 4.03AAFTEE502 pKa = 4.71YY503 pKa = 9.77TQPIVVNQAGEE514 pKa = 3.93HH515 pKa = 4.3TVRR518 pKa = 11.84YY519 pKa = 8.71RR520 pKa = 11.84ASDD523 pKa = 3.55NAGNTSDD530 pKa = 3.56VGSVAFTVVEE540 pKa = 4.19ADD542 pKa = 4.39PGDD545 pKa = 3.93SAPPEE550 pKa = 3.95VSAEE554 pKa = 4.14VGGEE558 pKa = 3.69TDD560 pKa = 3.1GQGNYY565 pKa = 9.72VGSATVTISASDD577 pKa = 3.76ADD579 pKa = 3.92SGIAAVEE586 pKa = 4.06YY587 pKa = 10.88ALDD590 pKa = 4.11DD591 pKa = 4.01AQFRR595 pKa = 11.84LYY597 pKa = 10.04EE598 pKa = 4.28GPVTVDD604 pKa = 2.86RR605 pKa = 11.84PGEE608 pKa = 3.84HH609 pKa = 4.94TVRR612 pKa = 11.84YY613 pKa = 9.08RR614 pKa = 11.84AVDD617 pKa = 3.38NAEE620 pKa = 3.91NTAEE624 pKa = 4.27GAVSFTVVADD634 pKa = 3.84PTDD637 pKa = 3.67ACPDD641 pKa = 3.32SDD643 pKa = 4.03EE644 pKa = 5.3RR645 pKa = 11.84PTVTIGDD652 pKa = 4.44DD653 pKa = 3.52DD654 pKa = 4.26TGVGNVDD661 pKa = 3.51TGNGCTINDD670 pKa = 5.45LIAEE674 pKa = 4.34DD675 pKa = 4.52AEE677 pKa = 4.56YY678 pKa = 11.09ANHH681 pKa = 6.97GEE683 pKa = 4.4FVDD686 pKa = 4.38HH687 pKa = 5.87VQEE690 pKa = 4.06LTRR693 pKa = 11.84EE694 pKa = 4.01LVEE697 pKa = 4.91AGIITDD703 pKa = 3.65GDD705 pKa = 3.42RR706 pKa = 11.84GRR708 pKa = 11.84IVSAAARR715 pKa = 11.84SDD717 pKa = 3.37VGKK720 pKa = 10.72
MM1 pKa = 7.45YY2 pKa = 10.59SRR4 pKa = 11.84LPSATPRR11 pKa = 11.84KK12 pKa = 8.49PRR14 pKa = 11.84PIARR18 pKa = 11.84LRR20 pKa = 11.84AAAGRR25 pKa = 11.84FAVAILGLLVATLWLAPASAAPRR48 pKa = 11.84TPEE51 pKa = 3.95PAAGTAQAQVLTWTANDD68 pKa = 4.45SITSYY73 pKa = 11.52ASAPSTATAGAATIVFEE90 pKa = 4.62NSTATGNTTGLPHH103 pKa = 7.24TLTFDD108 pKa = 3.43TSTPGYY114 pKa = 9.95NHH116 pKa = 7.1DD117 pKa = 3.5VDD119 pKa = 4.74VNILASPFDD128 pKa = 4.28ANEE131 pKa = 3.25GRR133 pKa = 11.84YY134 pKa = 8.55EE135 pKa = 4.18VNVNLSPGEE144 pKa = 3.83YY145 pKa = 9.14RR146 pKa = 11.84YY147 pKa = 10.47FCDD150 pKa = 3.89MPGHH154 pKa = 5.67GQMTGVLVVTGEE166 pKa = 4.37GSDD169 pKa = 3.39TTAPEE174 pKa = 3.79VSAEE178 pKa = 4.13VTGQTDD184 pKa = 3.25DD185 pKa = 3.13QGNYY189 pKa = 9.69LGSATVSVTATDD201 pKa = 3.39AGSGVASVEE210 pKa = 3.93YY211 pKa = 10.35EE212 pKa = 3.64IDD214 pKa = 3.43DD215 pKa = 4.21TGFQPYY221 pKa = 8.77TGPVTVDD228 pKa = 2.75RR229 pKa = 11.84VGDD232 pKa = 3.52HH233 pKa = 5.81SVQYY237 pKa = 10.47RR238 pKa = 11.84ATDD241 pKa = 3.44NAGNTSPTGSVQFSVVEE258 pKa = 4.12PDD260 pKa = 4.2PGDD263 pKa = 3.48TTPPEE268 pKa = 3.96VSAEE272 pKa = 4.16VGGEE276 pKa = 3.69TDD278 pKa = 3.1GQGNYY283 pKa = 9.72VGSATVTVAATDD295 pKa = 3.55TGSGVASVEE304 pKa = 4.06YY305 pKa = 9.95EE306 pKa = 3.62LDD308 pKa = 3.49GGGFRR313 pKa = 11.84TYY315 pKa = 10.45SAPVVVDD322 pKa = 3.11TEE324 pKa = 4.52GEE326 pKa = 4.12HH327 pKa = 4.82VVRR330 pKa = 11.84YY331 pKa = 9.11RR332 pKa = 11.84ATDD335 pKa = 3.18NAGNVSQSGSVSFTVVPPDD354 pKa = 4.55PGDD357 pKa = 3.45TTPPQVTAEE366 pKa = 4.0IAGEE370 pKa = 3.8RR371 pKa = 11.84DD372 pKa = 2.8AGGNYY377 pKa = 9.87LGSATVTLAAQDD389 pKa = 4.02SEE391 pKa = 4.87SQVDD395 pKa = 4.03SIEE398 pKa = 4.0YY399 pKa = 10.38AIGDD403 pKa = 4.06GAFQAYY409 pKa = 8.09AAPVTFDD416 pKa = 3.15QPGEE420 pKa = 3.82YY421 pKa = 9.05TLRR424 pKa = 11.84YY425 pKa = 9.21RR426 pKa = 11.84ATDD429 pKa = 3.29TAGNTSQPGSVSFAVVEE446 pKa = 4.34PPPDD450 pKa = 3.61DD451 pKa = 3.41TTAPEE456 pKa = 3.99VSAEE460 pKa = 4.15VGGEE464 pKa = 3.69TDD466 pKa = 3.1GQGNYY471 pKa = 9.72VGSATVTISASDD483 pKa = 3.76ADD485 pKa = 3.92SGIAAVEE492 pKa = 4.06YY493 pKa = 10.82ALDD496 pKa = 3.95DD497 pKa = 4.03AAFTEE502 pKa = 4.71YY503 pKa = 9.77TQPIVVNQAGEE514 pKa = 3.93HH515 pKa = 4.3TVRR518 pKa = 11.84YY519 pKa = 8.71RR520 pKa = 11.84ASDD523 pKa = 3.55NAGNTSDD530 pKa = 3.56VGSVAFTVVEE540 pKa = 4.19ADD542 pKa = 4.39PGDD545 pKa = 3.93SAPPEE550 pKa = 3.95VSAEE554 pKa = 4.14VGGEE558 pKa = 3.69TDD560 pKa = 3.1GQGNYY565 pKa = 9.72VGSATVTISASDD577 pKa = 3.76ADD579 pKa = 3.92SGIAAVEE586 pKa = 4.06YY587 pKa = 10.88ALDD590 pKa = 4.11DD591 pKa = 4.01AQFRR595 pKa = 11.84LYY597 pKa = 10.04EE598 pKa = 4.28GPVTVDD604 pKa = 2.86RR605 pKa = 11.84PGEE608 pKa = 3.84HH609 pKa = 4.94TVRR612 pKa = 11.84YY613 pKa = 9.08RR614 pKa = 11.84AVDD617 pKa = 3.38NAEE620 pKa = 3.91NTAEE624 pKa = 4.27GAVSFTVVADD634 pKa = 3.84PTDD637 pKa = 3.67ACPDD641 pKa = 3.32SDD643 pKa = 4.03EE644 pKa = 5.3RR645 pKa = 11.84PTVTIGDD652 pKa = 4.44DD653 pKa = 3.52DD654 pKa = 4.26TGVGNVDD661 pKa = 3.51TGNGCTINDD670 pKa = 5.45LIAEE674 pKa = 4.34DD675 pKa = 4.52AEE677 pKa = 4.56YY678 pKa = 11.09ANHH681 pKa = 6.97GEE683 pKa = 4.4FVDD686 pKa = 4.38HH687 pKa = 5.87VQEE690 pKa = 4.06LTRR693 pKa = 11.84EE694 pKa = 4.01LVEE697 pKa = 4.91AGIITDD703 pKa = 3.65GDD705 pKa = 3.42RR706 pKa = 11.84GRR708 pKa = 11.84IVSAAARR715 pKa = 11.84SDD717 pKa = 3.37VGKK720 pKa = 10.72
Molecular weight: 73.87 kDa
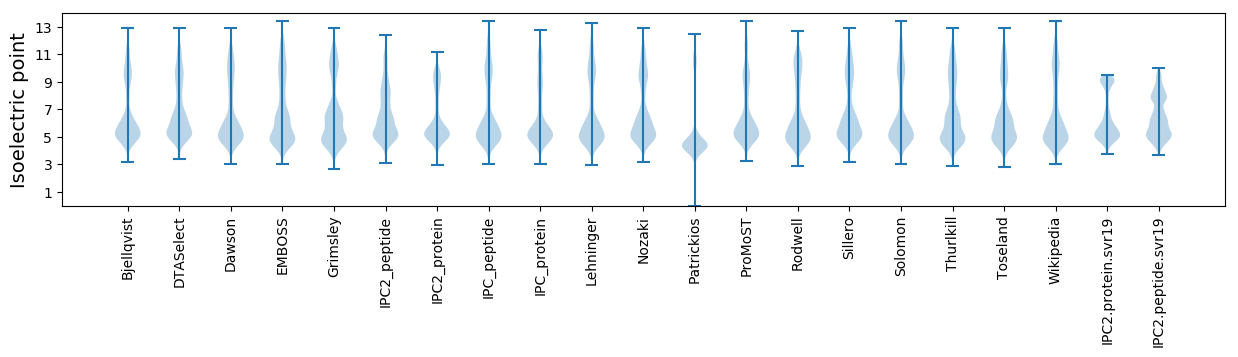
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|H5X4B9|H5X4B9_9PSEU Geranylgeranyl reductase family protein OS=Saccharomonospora marina XMU15 OX=882083 GN=SacmaDRAFT_0561 PE=4 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.38KK7 pKa = 8.42RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.06RR11 pKa = 11.84MSKK14 pKa = 9.76KK15 pKa = 9.54KK16 pKa = 9.72HH17 pKa = 5.63RR18 pKa = 11.84KK19 pKa = 7.56LLRR22 pKa = 11.84RR23 pKa = 11.84TRR25 pKa = 11.84VQRR28 pKa = 11.84RR29 pKa = 11.84KK30 pKa = 9.71QGKK33 pKa = 8.65
MM1 pKa = 7.4GSVIKK6 pKa = 10.38KK7 pKa = 8.42RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.06RR11 pKa = 11.84MSKK14 pKa = 9.76KK15 pKa = 9.54KK16 pKa = 9.72HH17 pKa = 5.63RR18 pKa = 11.84KK19 pKa = 7.56LLRR22 pKa = 11.84RR23 pKa = 11.84TRR25 pKa = 11.84VQRR28 pKa = 11.84RR29 pKa = 11.84KK30 pKa = 9.71QGKK33 pKa = 8.65
Molecular weight: 4.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1777747 |
25 |
14511 |
319.3 |
34.35 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.09 ± 0.044 | 0.798 ± 0.01 |
5.874 ± 0.022 | 5.974 ± 0.033 |
2.791 ± 0.02 | 9.126 ± 0.028 |
2.256 ± 0.014 | 3.344 ± 0.022 |
1.949 ± 0.023 | 10.581 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.72 ± 0.012 | 1.86 ± 0.017 |
5.84 ± 0.028 | 2.954 ± 0.022 |
8.376 ± 0.035 | 5.228 ± 0.022 |
5.849 ± 0.027 | 8.904 ± 0.034 |
1.487 ± 0.014 | 1.997 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
