
Groundnut rosette virus (strain MC1) (GRV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Tolucaviricetes; Tolivirales; Tombusviridae; Calvusvirinae; Umbravirus; Groundnut rosette virus
Average proteome isoelectric point is 7.61
Get precalculated fractions of proteins
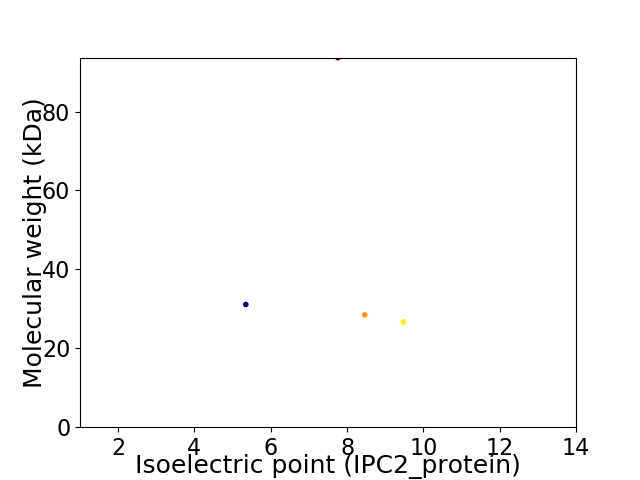
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q67683-2|RDRP-2_GRVMC Isoform of Q67683 Isoform ORF1 protein of RNA-directed RNA polymerase OS=Groundnut rosette virus (strain MC1) OX=1005060 GN=ORF1-ORF2 PE=4 SV=2
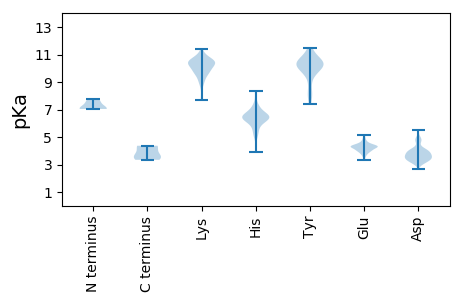
MM1 pKa = 7.04ATIRR5 pKa = 11.84HH6 pKa = 5.28IMDD9 pKa = 3.88WLRR12 pKa = 11.84PTFTPLAGVKK22 pKa = 9.61SRR24 pKa = 11.84QEE26 pKa = 4.29CIAHH30 pKa = 6.5YY31 pKa = 10.68GDD33 pKa = 4.92DD34 pKa = 3.84WALMITQSRR43 pKa = 11.84MTLSAEE49 pKa = 3.96AQVNAWYY56 pKa = 10.21EE57 pKa = 3.93GGAEE61 pKa = 4.28VNGSIPSVEE70 pKa = 4.2GNAAAPQAAAVVAEE84 pKa = 4.78PPHH87 pKa = 6.51PSPAEE92 pKa = 3.93NEE94 pKa = 4.37GEE96 pKa = 4.12DD97 pKa = 4.09ASWVAALPAPPTYY110 pKa = 10.48EE111 pKa = 4.2VVQGRR116 pKa = 11.84APTLEE121 pKa = 4.6EE122 pKa = 3.32IHH124 pKa = 6.19GASRR128 pKa = 11.84LQIVPYY134 pKa = 8.91TGRR137 pKa = 11.84ARR139 pKa = 11.84VIGEE143 pKa = 4.35DD144 pKa = 3.51EE145 pKa = 4.34VLPSPVLSSIWRR157 pKa = 11.84ATRR160 pKa = 11.84CPGRR164 pKa = 11.84ILRR167 pKa = 11.84VLGTLLKK174 pKa = 10.56PGACQRR180 pKa = 11.84HH181 pKa = 5.67LEE183 pKa = 4.31DD184 pKa = 4.17LRR186 pKa = 11.84EE187 pKa = 4.02VQPQVCVGNPCEE199 pKa = 4.45VKK201 pKa = 10.47LQEE204 pKa = 4.52EE205 pKa = 4.97GAPMAYY211 pKa = 9.6MNAQSIAMEE220 pKa = 4.32LRR222 pKa = 11.84AMFGWQAATPANRR235 pKa = 11.84EE236 pKa = 3.78LGNRR240 pKa = 11.84VARR243 pKa = 11.84DD244 pKa = 3.49ILRR247 pKa = 11.84DD248 pKa = 3.38GCGATRR254 pKa = 11.84EE255 pKa = 4.4QIWYY259 pKa = 7.9MSSLALHH266 pKa = 6.46MWFQPTLCDD275 pKa = 3.68LAIKK279 pKa = 10.59AGAQNFF285 pKa = 3.66
MM1 pKa = 7.04ATIRR5 pKa = 11.84HH6 pKa = 5.28IMDD9 pKa = 3.88WLRR12 pKa = 11.84PTFTPLAGVKK22 pKa = 9.61SRR24 pKa = 11.84QEE26 pKa = 4.29CIAHH30 pKa = 6.5YY31 pKa = 10.68GDD33 pKa = 4.92DD34 pKa = 3.84WALMITQSRR43 pKa = 11.84MTLSAEE49 pKa = 3.96AQVNAWYY56 pKa = 10.21EE57 pKa = 3.93GGAEE61 pKa = 4.28VNGSIPSVEE70 pKa = 4.2GNAAAPQAAAVVAEE84 pKa = 4.78PPHH87 pKa = 6.51PSPAEE92 pKa = 3.93NEE94 pKa = 4.37GEE96 pKa = 4.12DD97 pKa = 4.09ASWVAALPAPPTYY110 pKa = 10.48EE111 pKa = 4.2VVQGRR116 pKa = 11.84APTLEE121 pKa = 4.6EE122 pKa = 3.32IHH124 pKa = 6.19GASRR128 pKa = 11.84LQIVPYY134 pKa = 8.91TGRR137 pKa = 11.84ARR139 pKa = 11.84VIGEE143 pKa = 4.35DD144 pKa = 3.51EE145 pKa = 4.34VLPSPVLSSIWRR157 pKa = 11.84ATRR160 pKa = 11.84CPGRR164 pKa = 11.84ILRR167 pKa = 11.84VLGTLLKK174 pKa = 10.56PGACQRR180 pKa = 11.84HH181 pKa = 5.67LEE183 pKa = 4.31DD184 pKa = 4.17LRR186 pKa = 11.84EE187 pKa = 4.02VQPQVCVGNPCEE199 pKa = 4.45VKK201 pKa = 10.47LQEE204 pKa = 4.52EE205 pKa = 4.97GAPMAYY211 pKa = 9.6MNAQSIAMEE220 pKa = 4.32LRR222 pKa = 11.84AMFGWQAATPANRR235 pKa = 11.84EE236 pKa = 3.78LGNRR240 pKa = 11.84VARR243 pKa = 11.84DD244 pKa = 3.49ILRR247 pKa = 11.84DD248 pKa = 3.38GCGATRR254 pKa = 11.84EE255 pKa = 4.4QIWYY259 pKa = 7.9MSSLALHH266 pKa = 6.46MWFQPTLCDD275 pKa = 3.68LAIKK279 pKa = 10.59AGAQNFF285 pKa = 3.66
Molecular weight: 31.1 kDa
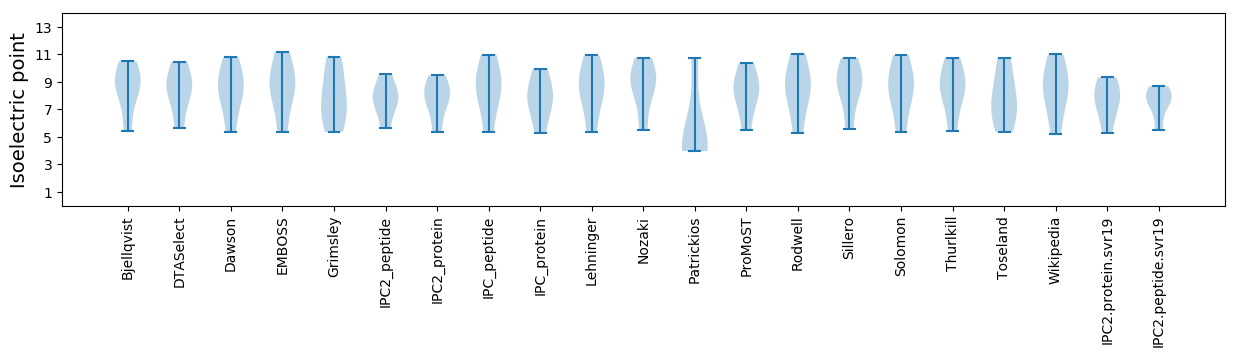
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q67685|MVP_GRVMC Movement protein OS=Groundnut rosette virus (strain MC1) OX=1005060 GN=ORF4 PE=1 SV=1
MM1 pKa = 7.76DD2 pKa = 4.09TTPASRR8 pKa = 11.84GQWPVGIMSSVINVFASGKK27 pKa = 8.87SCNSGGAPRR36 pKa = 11.84SSVRR40 pKa = 11.84RR41 pKa = 11.84GHH43 pKa = 6.45RR44 pKa = 11.84AGAARR49 pKa = 11.84DD50 pKa = 3.5KK51 pKa = 11.39SRR53 pKa = 11.84GFDD56 pKa = 3.26APPRR60 pKa = 11.84RR61 pKa = 11.84PKK63 pKa = 10.93GGVHH67 pKa = 7.12PATTSKK73 pKa = 11.16DD74 pKa = 3.44PNKK77 pKa = 10.34DD78 pKa = 2.68IRR80 pKa = 11.84GPPAPTPHH88 pKa = 6.57KK89 pKa = 10.0KK90 pKa = 10.13YY91 pKa = 10.01RR92 pKa = 11.84GPAIPGEE99 pKa = 4.39GRR101 pKa = 11.84SGVHH105 pKa = 3.89TTRR108 pKa = 11.84PRR110 pKa = 11.84RR111 pKa = 11.84RR112 pKa = 11.84AGRR115 pKa = 11.84SGGMDD120 pKa = 3.14PRR122 pKa = 11.84QLVAQPQQRR131 pKa = 11.84WAKK134 pKa = 9.37TEE136 pKa = 3.67IPTEE140 pKa = 3.79RR141 pKa = 11.84RR142 pKa = 11.84AEE144 pKa = 3.96IDD146 pKa = 3.33GLLPSLLNTLDD157 pKa = 3.64GQIQGDD163 pKa = 3.82AALLRR168 pKa = 11.84YY169 pKa = 9.39CVGAIKK175 pKa = 10.57RR176 pKa = 11.84EE177 pKa = 3.89LRR179 pKa = 11.84RR180 pKa = 11.84RR181 pKa = 11.84WEE183 pKa = 4.32SVQPAHH189 pKa = 6.63HH190 pKa = 6.39VAASSGKK197 pKa = 9.51PSPQLFNEE205 pKa = 4.76AAQNAEE211 pKa = 4.45DD212 pKa = 4.76LSGDD216 pKa = 3.66GKK218 pKa = 11.18GCAGQSVQQEE228 pKa = 4.46VLHH231 pKa = 6.55SGSGVPPVCADD242 pKa = 3.55CGKK245 pKa = 9.63PAANKK250 pKa = 8.04WW251 pKa = 3.34
MM1 pKa = 7.76DD2 pKa = 4.09TTPASRR8 pKa = 11.84GQWPVGIMSSVINVFASGKK27 pKa = 8.87SCNSGGAPRR36 pKa = 11.84SSVRR40 pKa = 11.84RR41 pKa = 11.84GHH43 pKa = 6.45RR44 pKa = 11.84AGAARR49 pKa = 11.84DD50 pKa = 3.5KK51 pKa = 11.39SRR53 pKa = 11.84GFDD56 pKa = 3.26APPRR60 pKa = 11.84RR61 pKa = 11.84PKK63 pKa = 10.93GGVHH67 pKa = 7.12PATTSKK73 pKa = 11.16DD74 pKa = 3.44PNKK77 pKa = 10.34DD78 pKa = 2.68IRR80 pKa = 11.84GPPAPTPHH88 pKa = 6.57KK89 pKa = 10.0KK90 pKa = 10.13YY91 pKa = 10.01RR92 pKa = 11.84GPAIPGEE99 pKa = 4.39GRR101 pKa = 11.84SGVHH105 pKa = 3.89TTRR108 pKa = 11.84PRR110 pKa = 11.84RR111 pKa = 11.84RR112 pKa = 11.84AGRR115 pKa = 11.84SGGMDD120 pKa = 3.14PRR122 pKa = 11.84QLVAQPQQRR131 pKa = 11.84WAKK134 pKa = 9.37TEE136 pKa = 3.67IPTEE140 pKa = 3.79RR141 pKa = 11.84RR142 pKa = 11.84AEE144 pKa = 3.96IDD146 pKa = 3.33GLLPSLLNTLDD157 pKa = 3.64GQIQGDD163 pKa = 3.82AALLRR168 pKa = 11.84YY169 pKa = 9.39CVGAIKK175 pKa = 10.57RR176 pKa = 11.84EE177 pKa = 3.89LRR179 pKa = 11.84RR180 pKa = 11.84RR181 pKa = 11.84WEE183 pKa = 4.32SVQPAHH189 pKa = 6.63HH190 pKa = 6.39VAASSGKK197 pKa = 9.51PSPQLFNEE205 pKa = 4.76AAQNAEE211 pKa = 4.45DD212 pKa = 4.76LSGDD216 pKa = 3.66GKK218 pKa = 11.18GCAGQSVQQEE228 pKa = 4.46VLHH231 pKa = 6.55SGSGVPPVCADD242 pKa = 3.55CGKK245 pKa = 9.63PAANKK250 pKa = 8.04WW251 pKa = 3.34
Molecular weight: 26.68 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
2474 |
251 |
839 |
494.8 |
54.71 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.065 ± 0.566 | 2.425 ± 0.306 |
4.365 ± 0.346 | 5.618 ± 0.366 |
2.587 ± 0.285 | 7.801 ± 0.546 |
2.264 ± 0.118 | 3.961 ± 0.223 |
4.244 ± 0.483 | 8.084 ± 0.487 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.112 ± 0.256 | 3.678 ± 0.28 |
7.114 ± 0.725 | 4.163 ± 0.215 |
7.316 ± 0.406 | 5.821 ± 0.595 |
4.325 ± 0.169 | 7.801 ± 0.343 |
2.061 ± 0.19 | 3.193 ± 0.449 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
