
Beihai sobemo-like virus 2
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
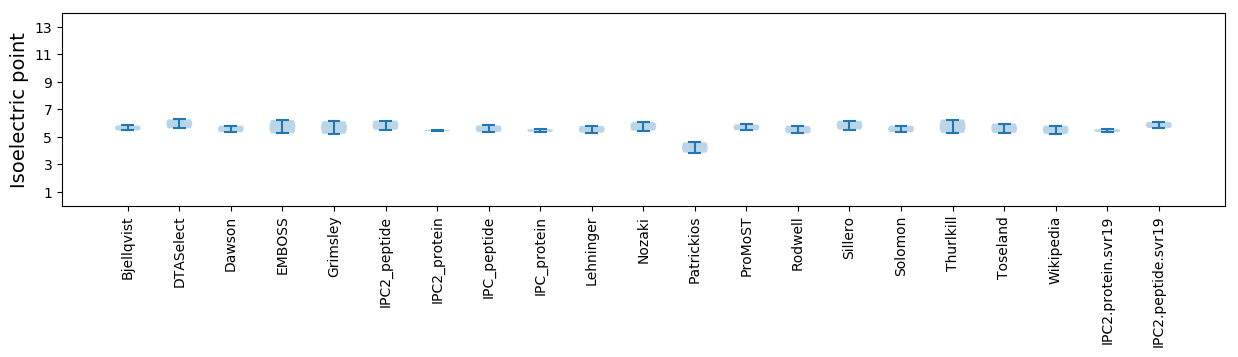
Average proteome isoelectric point is 5.46
Get precalculated fractions of proteins
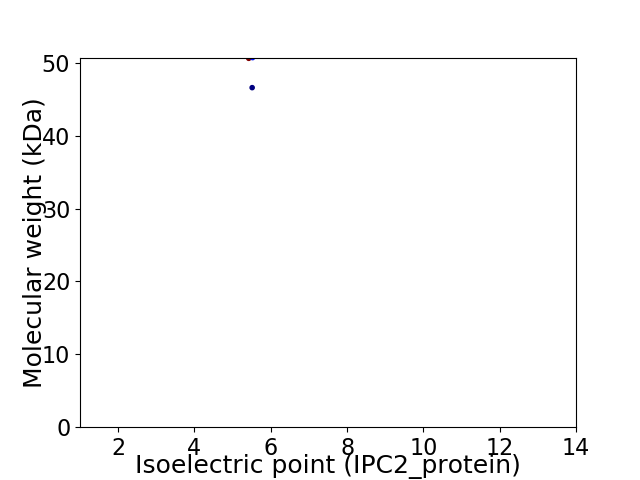
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
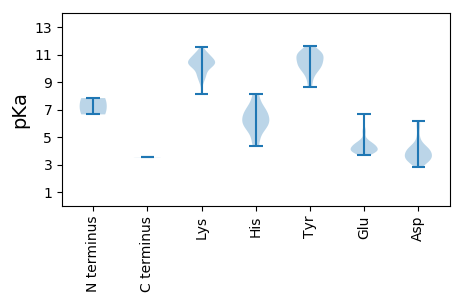
Protein with the lowest isoelectric point:
>tr|A0A1L3KE82|A0A1L3KE82_9VIRU Uncharacterized protein OS=Beihai sobemo-like virus 2 OX=1922691 PE=4 SV=1
MM1 pKa = 6.65STKK4 pKa = 10.49DD5 pKa = 3.33KK6 pKa = 10.87RR7 pKa = 11.84IVDD10 pKa = 3.55DD11 pKa = 3.84TVRR14 pKa = 11.84EE15 pKa = 3.87LSKK18 pKa = 10.51RR19 pKa = 11.84SKK21 pKa = 10.13PNKK24 pKa = 8.16KK25 pKa = 10.2SKK27 pKa = 10.73VEE29 pKa = 4.36SIIQTAGSYY38 pKa = 10.42DD39 pKa = 3.54LQYY42 pKa = 10.96FDD44 pKa = 5.79EE45 pKa = 5.36ISLDD49 pKa = 3.53FLKK52 pKa = 10.77YY53 pKa = 9.63RR54 pKa = 11.84HH55 pKa = 5.82QVDD58 pKa = 3.68KK59 pKa = 9.03PTSSFEE65 pKa = 3.85TWKK68 pKa = 11.01DD69 pKa = 3.31SLTPPVKK76 pKa = 10.37VSMMKK81 pKa = 9.53EE82 pKa = 3.86WKK84 pKa = 9.57KK85 pKa = 11.12KK86 pKa = 9.68QMDD89 pKa = 4.43RR90 pKa = 11.84NWEE93 pKa = 4.19DD94 pKa = 3.68DD95 pKa = 4.03PVPGWRR101 pKa = 11.84VPDD104 pKa = 4.13ALQKK108 pKa = 10.71KK109 pKa = 9.94YY110 pKa = 10.89GFEE113 pKa = 4.13FLRR116 pKa = 11.84ATPTAFSVPLEE127 pKa = 4.44DD128 pKa = 4.54SNEE131 pKa = 4.03VVQKK135 pKa = 10.92AIIDD139 pKa = 3.94TPPSYY144 pKa = 10.59RR145 pKa = 11.84PPTEE149 pKa = 5.73DD150 pKa = 3.02IFGDD154 pKa = 3.7DD155 pKa = 3.55EE156 pKa = 4.8VKK158 pKa = 11.05NEE160 pKa = 4.21ADD162 pKa = 4.08DD163 pKa = 4.64PPAEE167 pKa = 4.43SGSDD171 pKa = 3.31GMNEE175 pKa = 3.85PTEE178 pKa = 4.4TGPSNPGVKK187 pKa = 9.23SVYY190 pKa = 10.14IVDD193 pKa = 3.86YY194 pKa = 11.15DD195 pKa = 4.8DD196 pKa = 6.03DD197 pKa = 4.24KK198 pKa = 11.58QLPTEE203 pKa = 4.5EE204 pKa = 4.21IVDD207 pKa = 4.14IINNYY212 pKa = 8.67VVMNVAYY219 pKa = 10.24NAYY222 pKa = 10.16FEE224 pKa = 5.41DD225 pKa = 5.29AIEE228 pKa = 5.24DD229 pKa = 3.48IRR231 pKa = 11.84NGDD234 pKa = 3.39KK235 pKa = 10.61MKK237 pKa = 10.94GRR239 pKa = 11.84TLEE242 pKa = 3.97KK243 pKa = 10.23LQNRR247 pKa = 11.84LAGAWGALKK256 pKa = 10.57ALRR259 pKa = 11.84KK260 pKa = 8.92QSNYY264 pKa = 9.99HH265 pKa = 6.36IAVQLANYY273 pKa = 9.38KK274 pKa = 10.36GVEE277 pKa = 4.01ALHH280 pKa = 8.05SEE282 pKa = 4.34MNQVTITMAEE292 pKa = 3.69RR293 pKa = 11.84SEE295 pKa = 4.32IIARR299 pKa = 11.84IYY301 pKa = 11.12DD302 pKa = 3.63DD303 pKa = 6.14AITKK307 pKa = 10.49SKK309 pKa = 9.41VQEE312 pKa = 4.63RR313 pKa = 11.84IDD315 pKa = 4.19KK316 pKa = 9.9IAKK319 pKa = 9.0EE320 pKa = 3.95LSEE323 pKa = 4.59LRR325 pKa = 11.84DD326 pKa = 3.84NIQSCKK332 pKa = 9.84EE333 pKa = 3.73IKK335 pKa = 10.51NPSSKK340 pKa = 10.82LDD342 pKa = 3.61LLASVEE348 pKa = 4.35SKK350 pKa = 10.36IPKK353 pKa = 9.49IKK355 pKa = 10.53PEE357 pKa = 3.97ADD359 pKa = 3.22YY360 pKa = 10.51MPNEE364 pKa = 4.16KK365 pKa = 10.32SGLAAEE371 pKa = 4.5LEE373 pKa = 4.48KK374 pKa = 10.93PFNEE378 pKa = 4.64AMNQEE383 pKa = 4.13IEE385 pKa = 4.33KK386 pKa = 10.59KK387 pKa = 10.34ALEE390 pKa = 4.21LLEE393 pKa = 4.69SVQSLNLVAPLGLKK407 pKa = 10.7DD408 pKa = 3.7MLRR411 pKa = 11.84GATSSSSSMITPEE424 pKa = 3.77SSTKK428 pKa = 10.26KK429 pKa = 10.13KK430 pKa = 9.26RR431 pKa = 11.84TRR433 pKa = 11.84SKK435 pKa = 10.42KK436 pKa = 10.22KK437 pKa = 10.25KK438 pKa = 10.5VGDD441 pKa = 3.92GSDD444 pKa = 4.09KK445 pKa = 10.77RR446 pKa = 11.84DD447 pKa = 3.27QQ448 pKa = 3.51
MM1 pKa = 6.65STKK4 pKa = 10.49DD5 pKa = 3.33KK6 pKa = 10.87RR7 pKa = 11.84IVDD10 pKa = 3.55DD11 pKa = 3.84TVRR14 pKa = 11.84EE15 pKa = 3.87LSKK18 pKa = 10.51RR19 pKa = 11.84SKK21 pKa = 10.13PNKK24 pKa = 8.16KK25 pKa = 10.2SKK27 pKa = 10.73VEE29 pKa = 4.36SIIQTAGSYY38 pKa = 10.42DD39 pKa = 3.54LQYY42 pKa = 10.96FDD44 pKa = 5.79EE45 pKa = 5.36ISLDD49 pKa = 3.53FLKK52 pKa = 10.77YY53 pKa = 9.63RR54 pKa = 11.84HH55 pKa = 5.82QVDD58 pKa = 3.68KK59 pKa = 9.03PTSSFEE65 pKa = 3.85TWKK68 pKa = 11.01DD69 pKa = 3.31SLTPPVKK76 pKa = 10.37VSMMKK81 pKa = 9.53EE82 pKa = 3.86WKK84 pKa = 9.57KK85 pKa = 11.12KK86 pKa = 9.68QMDD89 pKa = 4.43RR90 pKa = 11.84NWEE93 pKa = 4.19DD94 pKa = 3.68DD95 pKa = 4.03PVPGWRR101 pKa = 11.84VPDD104 pKa = 4.13ALQKK108 pKa = 10.71KK109 pKa = 9.94YY110 pKa = 10.89GFEE113 pKa = 4.13FLRR116 pKa = 11.84ATPTAFSVPLEE127 pKa = 4.44DD128 pKa = 4.54SNEE131 pKa = 4.03VVQKK135 pKa = 10.92AIIDD139 pKa = 3.94TPPSYY144 pKa = 10.59RR145 pKa = 11.84PPTEE149 pKa = 5.73DD150 pKa = 3.02IFGDD154 pKa = 3.7DD155 pKa = 3.55EE156 pKa = 4.8VKK158 pKa = 11.05NEE160 pKa = 4.21ADD162 pKa = 4.08DD163 pKa = 4.64PPAEE167 pKa = 4.43SGSDD171 pKa = 3.31GMNEE175 pKa = 3.85PTEE178 pKa = 4.4TGPSNPGVKK187 pKa = 9.23SVYY190 pKa = 10.14IVDD193 pKa = 3.86YY194 pKa = 11.15DD195 pKa = 4.8DD196 pKa = 6.03DD197 pKa = 4.24KK198 pKa = 11.58QLPTEE203 pKa = 4.5EE204 pKa = 4.21IVDD207 pKa = 4.14IINNYY212 pKa = 8.67VVMNVAYY219 pKa = 10.24NAYY222 pKa = 10.16FEE224 pKa = 5.41DD225 pKa = 5.29AIEE228 pKa = 5.24DD229 pKa = 3.48IRR231 pKa = 11.84NGDD234 pKa = 3.39KK235 pKa = 10.61MKK237 pKa = 10.94GRR239 pKa = 11.84TLEE242 pKa = 3.97KK243 pKa = 10.23LQNRR247 pKa = 11.84LAGAWGALKK256 pKa = 10.57ALRR259 pKa = 11.84KK260 pKa = 8.92QSNYY264 pKa = 9.99HH265 pKa = 6.36IAVQLANYY273 pKa = 9.38KK274 pKa = 10.36GVEE277 pKa = 4.01ALHH280 pKa = 8.05SEE282 pKa = 4.34MNQVTITMAEE292 pKa = 3.69RR293 pKa = 11.84SEE295 pKa = 4.32IIARR299 pKa = 11.84IYY301 pKa = 11.12DD302 pKa = 3.63DD303 pKa = 6.14AITKK307 pKa = 10.49SKK309 pKa = 9.41VQEE312 pKa = 4.63RR313 pKa = 11.84IDD315 pKa = 4.19KK316 pKa = 9.9IAKK319 pKa = 9.0EE320 pKa = 3.95LSEE323 pKa = 4.59LRR325 pKa = 11.84DD326 pKa = 3.84NIQSCKK332 pKa = 9.84EE333 pKa = 3.73IKK335 pKa = 10.51NPSSKK340 pKa = 10.82LDD342 pKa = 3.61LLASVEE348 pKa = 4.35SKK350 pKa = 10.36IPKK353 pKa = 9.49IKK355 pKa = 10.53PEE357 pKa = 3.97ADD359 pKa = 3.22YY360 pKa = 10.51MPNEE364 pKa = 4.16KK365 pKa = 10.32SGLAAEE371 pKa = 4.5LEE373 pKa = 4.48KK374 pKa = 10.93PFNEE378 pKa = 4.64AMNQEE383 pKa = 4.13IEE385 pKa = 4.33KK386 pKa = 10.59KK387 pKa = 10.34ALEE390 pKa = 4.21LLEE393 pKa = 4.69SVQSLNLVAPLGLKK407 pKa = 10.7DD408 pKa = 3.7MLRR411 pKa = 11.84GATSSSSSMITPEE424 pKa = 3.77SSTKK428 pKa = 10.26KK429 pKa = 10.13KK430 pKa = 9.26RR431 pKa = 11.84TRR433 pKa = 11.84SKK435 pKa = 10.42KK436 pKa = 10.22KK437 pKa = 10.25KK438 pKa = 10.5VGDD441 pKa = 3.92GSDD444 pKa = 4.09KK445 pKa = 10.77RR446 pKa = 11.84DD447 pKa = 3.27QQ448 pKa = 3.51
Molecular weight: 50.63 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KE82|A0A1L3KE82_9VIRU Uncharacterized protein OS=Beihai sobemo-like virus 2 OX=1922691 PE=4 SV=1
MM1 pKa = 7.83EE2 pKa = 6.7DD3 pKa = 4.42DD4 pKa = 3.79FTVTIPRR11 pKa = 11.84LWDD14 pKa = 3.21MMSAYY19 pKa = 10.18HH20 pKa = 6.77LSCNTKK26 pKa = 9.72AHH28 pKa = 6.77PGVPAYY34 pKa = 10.71VLADD38 pKa = 3.72EE39 pKa = 5.47KK40 pKa = 11.17DD41 pKa = 4.02TILKK45 pKa = 8.15THH47 pKa = 7.15LASVIKK53 pKa = 10.54AGFHH57 pKa = 5.03RR58 pKa = 11.84AVRR61 pKa = 11.84RR62 pKa = 11.84IAFYY66 pKa = 10.67YY67 pKa = 9.48VIRR70 pKa = 11.84EE71 pKa = 3.98NNYY74 pKa = 9.93DD75 pKa = 3.41YY76 pKa = 11.24HH77 pKa = 8.16FSAEE81 pKa = 3.94QLVVFGLCDD90 pKa = 4.96PIRR93 pKa = 11.84LMIKK97 pKa = 10.24DD98 pKa = 4.2EE99 pKa = 3.93PHH101 pKa = 5.98SLKK104 pKa = 10.66KK105 pKa = 10.26QLQGRR110 pKa = 11.84HH111 pKa = 4.54RR112 pKa = 11.84LIWVCSMFDD121 pKa = 3.39EE122 pKa = 4.72MADD125 pKa = 3.53KK126 pKa = 11.23AFLRR130 pKa = 11.84LDD132 pKa = 4.1KK133 pKa = 11.24LCIDD137 pKa = 3.53QWRR140 pKa = 11.84NIPMKK145 pKa = 10.09PGAPLGHH152 pKa = 5.69QQGDD156 pKa = 3.2ANMIQFMKK164 pKa = 10.47RR165 pKa = 11.84NPKK168 pKa = 10.08LKK170 pKa = 9.52STDD173 pKa = 2.82VSYY176 pKa = 11.12FDD178 pKa = 3.21WMYY181 pKa = 11.19NEE183 pKa = 4.5HH184 pKa = 6.66LHH186 pKa = 5.96RR187 pKa = 11.84CNIIARR193 pKa = 11.84FGLKK197 pKa = 10.14KK198 pKa = 10.46YY199 pKa = 10.59DD200 pKa = 3.73FNEE203 pKa = 3.97LEE205 pKa = 4.18RR206 pKa = 11.84LYY208 pKa = 11.09SNPLEE213 pKa = 4.28VDD215 pKa = 3.26DD216 pKa = 5.98FYY218 pKa = 11.24WAGLYY223 pKa = 8.88VQHH226 pKa = 6.93LCHH229 pKa = 5.79MRR231 pKa = 11.84KK232 pKa = 10.04VIVLSDD238 pKa = 3.11GNMYY242 pKa = 10.08GQQFTGINPSGNAGTTNHH260 pKa = 6.07NNFGRR265 pKa = 11.84LLVHH269 pKa = 7.22AEE271 pKa = 3.91ACVQSGVHH279 pKa = 6.73DD280 pKa = 4.23LQAVVFGDD288 pKa = 3.99DD289 pKa = 4.24CVTQYY294 pKa = 11.66GDD296 pKa = 3.49EE297 pKa = 4.75LKK299 pKa = 11.17DD300 pKa = 2.99EE301 pKa = 4.46DD302 pKa = 5.56AILRR306 pKa = 11.84EE307 pKa = 3.84YY308 pKa = 10.95GVIVKK313 pKa = 10.18EE314 pKa = 4.0EE315 pKa = 3.78EE316 pKa = 4.37TIISEE321 pKa = 4.04NNGRR325 pKa = 11.84HH326 pKa = 4.36TMSFCSRR333 pKa = 11.84RR334 pKa = 11.84YY335 pKa = 9.9DD336 pKa = 2.85MWMEE340 pKa = 4.49GDD342 pKa = 4.11QLMCTDD348 pKa = 3.99TFLNVDD354 pKa = 4.23KK355 pKa = 11.02LLSSYY360 pKa = 10.59LHH362 pKa = 6.19SNRR365 pKa = 11.84DD366 pKa = 3.35KK367 pKa = 10.49QHH369 pKa = 5.8QDD371 pKa = 3.0SVEE374 pKa = 3.98MEE376 pKa = 4.04LGIEE380 pKa = 4.05FVEE383 pKa = 4.23WLNSLEE389 pKa = 4.04RR390 pKa = 11.84STGGLEE396 pKa = 4.17GARR399 pKa = 11.84KK400 pKa = 9.45NLL402 pKa = 3.53
MM1 pKa = 7.83EE2 pKa = 6.7DD3 pKa = 4.42DD4 pKa = 3.79FTVTIPRR11 pKa = 11.84LWDD14 pKa = 3.21MMSAYY19 pKa = 10.18HH20 pKa = 6.77LSCNTKK26 pKa = 9.72AHH28 pKa = 6.77PGVPAYY34 pKa = 10.71VLADD38 pKa = 3.72EE39 pKa = 5.47KK40 pKa = 11.17DD41 pKa = 4.02TILKK45 pKa = 8.15THH47 pKa = 7.15LASVIKK53 pKa = 10.54AGFHH57 pKa = 5.03RR58 pKa = 11.84AVRR61 pKa = 11.84RR62 pKa = 11.84IAFYY66 pKa = 10.67YY67 pKa = 9.48VIRR70 pKa = 11.84EE71 pKa = 3.98NNYY74 pKa = 9.93DD75 pKa = 3.41YY76 pKa = 11.24HH77 pKa = 8.16FSAEE81 pKa = 3.94QLVVFGLCDD90 pKa = 4.96PIRR93 pKa = 11.84LMIKK97 pKa = 10.24DD98 pKa = 4.2EE99 pKa = 3.93PHH101 pKa = 5.98SLKK104 pKa = 10.66KK105 pKa = 10.26QLQGRR110 pKa = 11.84HH111 pKa = 4.54RR112 pKa = 11.84LIWVCSMFDD121 pKa = 3.39EE122 pKa = 4.72MADD125 pKa = 3.53KK126 pKa = 11.23AFLRR130 pKa = 11.84LDD132 pKa = 4.1KK133 pKa = 11.24LCIDD137 pKa = 3.53QWRR140 pKa = 11.84NIPMKK145 pKa = 10.09PGAPLGHH152 pKa = 5.69QQGDD156 pKa = 3.2ANMIQFMKK164 pKa = 10.47RR165 pKa = 11.84NPKK168 pKa = 10.08LKK170 pKa = 9.52STDD173 pKa = 2.82VSYY176 pKa = 11.12FDD178 pKa = 3.21WMYY181 pKa = 11.19NEE183 pKa = 4.5HH184 pKa = 6.66LHH186 pKa = 5.96RR187 pKa = 11.84CNIIARR193 pKa = 11.84FGLKK197 pKa = 10.14KK198 pKa = 10.46YY199 pKa = 10.59DD200 pKa = 3.73FNEE203 pKa = 3.97LEE205 pKa = 4.18RR206 pKa = 11.84LYY208 pKa = 11.09SNPLEE213 pKa = 4.28VDD215 pKa = 3.26DD216 pKa = 5.98FYY218 pKa = 11.24WAGLYY223 pKa = 8.88VQHH226 pKa = 6.93LCHH229 pKa = 5.79MRR231 pKa = 11.84KK232 pKa = 10.04VIVLSDD238 pKa = 3.11GNMYY242 pKa = 10.08GQQFTGINPSGNAGTTNHH260 pKa = 6.07NNFGRR265 pKa = 11.84LLVHH269 pKa = 7.22AEE271 pKa = 3.91ACVQSGVHH279 pKa = 6.73DD280 pKa = 4.23LQAVVFGDD288 pKa = 3.99DD289 pKa = 4.24CVTQYY294 pKa = 11.66GDD296 pKa = 3.49EE297 pKa = 4.75LKK299 pKa = 11.17DD300 pKa = 2.99EE301 pKa = 4.46DD302 pKa = 5.56AILRR306 pKa = 11.84EE307 pKa = 3.84YY308 pKa = 10.95GVIVKK313 pKa = 10.18EE314 pKa = 4.0EE315 pKa = 3.78EE316 pKa = 4.37TIISEE321 pKa = 4.04NNGRR325 pKa = 11.84HH326 pKa = 4.36TMSFCSRR333 pKa = 11.84RR334 pKa = 11.84YY335 pKa = 9.9DD336 pKa = 2.85MWMEE340 pKa = 4.49GDD342 pKa = 4.11QLMCTDD348 pKa = 3.99TFLNVDD354 pKa = 4.23KK355 pKa = 11.02LLSSYY360 pKa = 10.59LHH362 pKa = 6.19SNRR365 pKa = 11.84DD366 pKa = 3.35KK367 pKa = 10.49QHH369 pKa = 5.8QDD371 pKa = 3.0SVEE374 pKa = 3.98MEE376 pKa = 4.04LGIEE380 pKa = 4.05FVEE383 pKa = 4.23WLNSLEE389 pKa = 4.04RR390 pKa = 11.84STGGLEE396 pKa = 4.17GARR399 pKa = 11.84KK400 pKa = 9.45NLL402 pKa = 3.53
Molecular weight: 46.6 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
850 |
402 |
448 |
425.0 |
48.61 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.882 ± 0.42 | 1.294 ± 0.761 |
8.118 ± 0.259 | 7.529 ± 0.835 |
3.176 ± 0.83 | 5.059 ± 0.74 |
2.471 ± 1.28 | 5.529 ± 0.353 |
8.353 ± 1.995 | 8.235 ± 0.935 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.529 ± 0.446 | 5.059 ± 0.264 |
4.353 ± 1.031 | 3.765 ± 0.137 |
5.059 ± 0.422 | 7.176 ± 1.245 |
4.235 ± 0.321 | 6.118 ± 0.065 |
1.412 ± 0.21 | 3.647 ± 0.371 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
