
Eggerthella sp. (strain YY7918)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Coriobacteriia; Eggerthellales; Eggerthellaceae; Eggerthella; unclassified Eggerthella
Average proteome isoelectric point is 5.75
Get precalculated fractions of proteins
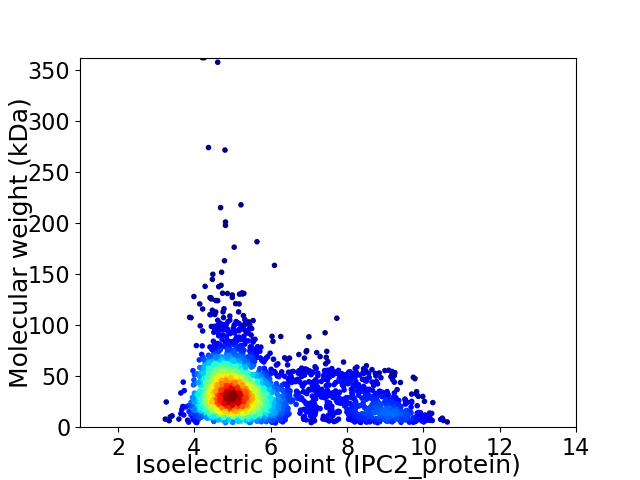
Virtual 2D-PAGE plot for 2674 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F7UZP0|F7UZP0_EEGSY Lysine--tRNA ligase OS=Eggerthella sp. (strain YY7918) OX=502558 GN=LysU PE=3 SV=1
MM1 pKa = 7.57LLASLIALGVIAYY14 pKa = 8.41NYY16 pKa = 8.18WQGQQLYY23 pKa = 11.0DD24 pKa = 4.56EE25 pKa = 5.16IADD28 pKa = 3.64QGFVPPTDD36 pKa = 3.35IEE38 pKa = 4.44GTALADD44 pKa = 3.61ITVDD48 pKa = 2.9WDD50 pKa = 3.41ALKK53 pKa = 10.68AINPDD58 pKa = 3.42TVGWIYY64 pKa = 10.7IPDD67 pKa = 3.8TQVNYY72 pKa = 9.92PIVHH76 pKa = 5.23TTDD79 pKa = 2.79NEE81 pKa = 4.36KK82 pKa = 11.04YY83 pKa = 8.96LTRR86 pKa = 11.84DD87 pKa = 3.47FKK89 pKa = 11.22GTEE92 pKa = 3.6GWLAQYY98 pKa = 10.7GAIFLAAEE106 pKa = 4.14NKK108 pKa = 10.56GDD110 pKa = 4.32FSDD113 pKa = 4.76SNNILYY119 pKa = 8.9GHH121 pKa = 6.37NMQDD125 pKa = 2.89GSMFACVSGFTDD137 pKa = 3.47ATQFAEE143 pKa = 3.85HH144 pKa = 5.9RR145 pKa = 11.84TIYY148 pKa = 11.0LLTPQGNYY156 pKa = 10.04RR157 pKa = 11.84LQTFALIHH165 pKa = 5.17TTADD169 pKa = 3.41DD170 pKa = 5.6LIAQTTFTDD179 pKa = 3.74EE180 pKa = 4.2EE181 pKa = 3.85QRR183 pKa = 11.84RR184 pKa = 11.84AYY186 pKa = 10.46LQDD189 pKa = 4.1KK190 pKa = 9.68IDD192 pKa = 3.84RR193 pKa = 11.84SVASVDD199 pKa = 3.52DD200 pKa = 4.19VPAVSEE206 pKa = 4.24MSQSLMLSTCDD217 pKa = 4.13NLPTDD222 pKa = 3.63GRR224 pKa = 11.84YY225 pKa = 9.6VLYY228 pKa = 10.46AYY230 pKa = 10.22VAEE233 pKa = 4.48STVKK237 pKa = 10.6ASSDD241 pKa = 3.25ASQGGAASPDD251 pKa = 3.29AVDD254 pKa = 5.18AVDD257 pKa = 3.65EE258 pKa = 4.41ASKK261 pKa = 10.88EE262 pKa = 4.06LASS265 pKa = 3.79
MM1 pKa = 7.57LLASLIALGVIAYY14 pKa = 8.41NYY16 pKa = 8.18WQGQQLYY23 pKa = 11.0DD24 pKa = 4.56EE25 pKa = 5.16IADD28 pKa = 3.64QGFVPPTDD36 pKa = 3.35IEE38 pKa = 4.44GTALADD44 pKa = 3.61ITVDD48 pKa = 2.9WDD50 pKa = 3.41ALKK53 pKa = 10.68AINPDD58 pKa = 3.42TVGWIYY64 pKa = 10.7IPDD67 pKa = 3.8TQVNYY72 pKa = 9.92PIVHH76 pKa = 5.23TTDD79 pKa = 2.79NEE81 pKa = 4.36KK82 pKa = 11.04YY83 pKa = 8.96LTRR86 pKa = 11.84DD87 pKa = 3.47FKK89 pKa = 11.22GTEE92 pKa = 3.6GWLAQYY98 pKa = 10.7GAIFLAAEE106 pKa = 4.14NKK108 pKa = 10.56GDD110 pKa = 4.32FSDD113 pKa = 4.76SNNILYY119 pKa = 8.9GHH121 pKa = 6.37NMQDD125 pKa = 2.89GSMFACVSGFTDD137 pKa = 3.47ATQFAEE143 pKa = 3.85HH144 pKa = 5.9RR145 pKa = 11.84TIYY148 pKa = 11.0LLTPQGNYY156 pKa = 10.04RR157 pKa = 11.84LQTFALIHH165 pKa = 5.17TTADD169 pKa = 3.41DD170 pKa = 5.6LIAQTTFTDD179 pKa = 3.74EE180 pKa = 4.2EE181 pKa = 3.85QRR183 pKa = 11.84RR184 pKa = 11.84AYY186 pKa = 10.46LQDD189 pKa = 4.1KK190 pKa = 9.68IDD192 pKa = 3.84RR193 pKa = 11.84SVASVDD199 pKa = 3.52DD200 pKa = 4.19VPAVSEE206 pKa = 4.24MSQSLMLSTCDD217 pKa = 4.13NLPTDD222 pKa = 3.63GRR224 pKa = 11.84YY225 pKa = 9.6VLYY228 pKa = 10.46AYY230 pKa = 10.22VAEE233 pKa = 4.48STVKK237 pKa = 10.6ASSDD241 pKa = 3.25ASQGGAASPDD251 pKa = 3.29AVDD254 pKa = 5.18AVDD257 pKa = 3.65EE258 pKa = 4.41ASKK261 pKa = 10.88EE262 pKa = 4.06LASS265 pKa = 3.79
Molecular weight: 29.05 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F7UZU6|F7UZU6_EEGSY Putative agmatine deiminase OS=Eggerthella sp. (strain YY7918) OX=502558 GN=aguA PE=3 SV=1
MM1 pKa = 7.06GAKK4 pKa = 9.37RR5 pKa = 11.84HH6 pKa = 4.96EE7 pKa = 4.08RR8 pKa = 11.84TRR10 pKa = 11.84ARR12 pKa = 11.84RR13 pKa = 11.84AAIQVLYY20 pKa = 9.69MGEE23 pKa = 3.84IRR25 pKa = 11.84EE26 pKa = 4.29LSPLTIAEE34 pKa = 4.42SDD36 pKa = 3.61DD37 pKa = 3.35SHH39 pKa = 8.56LIEE42 pKa = 5.24GGPIPVYY49 pKa = 10.62ALEE52 pKa = 4.24LVRR55 pKa = 11.84GVCEE59 pKa = 4.29HH60 pKa = 6.72PNEE63 pKa = 4.15VDD65 pKa = 3.27EE66 pKa = 5.81HH67 pKa = 5.65LTAPLKK73 pKa = 9.22TGRR76 pKa = 11.84WDD78 pKa = 3.57TLQNAWMARR87 pKa = 11.84HH88 pKa = 5.63GRR90 pKa = 11.84RR91 pKa = 11.84GRR93 pKa = 11.84CLRR96 pKa = 11.84SLRR99 pKa = 11.84YY100 pKa = 9.03FPRR103 pKa = 11.84AKK105 pKa = 10.46RR106 pKa = 11.84MLCAKK111 pKa = 9.94GMAWLFWRR119 pKa = 11.84LGAWRR124 pKa = 11.84RR125 pKa = 11.84ALGAAEE131 pKa = 4.33LLEE134 pKa = 4.33AQGVDD139 pKa = 3.02ARR141 pKa = 11.84VVDD144 pKa = 4.03MRR146 pKa = 11.84WVKK149 pKa = 10.31PLDD152 pKa = 3.75AEE154 pKa = 4.03EE155 pKa = 4.19MPRR158 pKa = 11.84VLLRR162 pKa = 11.84RR163 pKa = 11.84FEE165 pKa = 4.65STGSRR170 pKa = 11.84YY171 pKa = 10.23KK172 pKa = 10.44FWGRR176 pKa = 3.23
MM1 pKa = 7.06GAKK4 pKa = 9.37RR5 pKa = 11.84HH6 pKa = 4.96EE7 pKa = 4.08RR8 pKa = 11.84TRR10 pKa = 11.84ARR12 pKa = 11.84RR13 pKa = 11.84AAIQVLYY20 pKa = 9.69MGEE23 pKa = 3.84IRR25 pKa = 11.84EE26 pKa = 4.29LSPLTIAEE34 pKa = 4.42SDD36 pKa = 3.61DD37 pKa = 3.35SHH39 pKa = 8.56LIEE42 pKa = 5.24GGPIPVYY49 pKa = 10.62ALEE52 pKa = 4.24LVRR55 pKa = 11.84GVCEE59 pKa = 4.29HH60 pKa = 6.72PNEE63 pKa = 4.15VDD65 pKa = 3.27EE66 pKa = 5.81HH67 pKa = 5.65LTAPLKK73 pKa = 9.22TGRR76 pKa = 11.84WDD78 pKa = 3.57TLQNAWMARR87 pKa = 11.84HH88 pKa = 5.63GRR90 pKa = 11.84RR91 pKa = 11.84GRR93 pKa = 11.84CLRR96 pKa = 11.84SLRR99 pKa = 11.84YY100 pKa = 9.03FPRR103 pKa = 11.84AKK105 pKa = 10.46RR106 pKa = 11.84MLCAKK111 pKa = 9.94GMAWLFWRR119 pKa = 11.84LGAWRR124 pKa = 11.84RR125 pKa = 11.84ALGAAEE131 pKa = 4.33LLEE134 pKa = 4.33AQGVDD139 pKa = 3.02ARR141 pKa = 11.84VVDD144 pKa = 4.03MRR146 pKa = 11.84WVKK149 pKa = 10.31PLDD152 pKa = 3.75AEE154 pKa = 4.03EE155 pKa = 4.19MPRR158 pKa = 11.84VLLRR162 pKa = 11.84RR163 pKa = 11.84FEE165 pKa = 4.65STGSRR170 pKa = 11.84YY171 pKa = 10.23KK172 pKa = 10.44FWGRR176 pKa = 3.23
Molecular weight: 20.41 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
912144 |
39 |
3501 |
341.1 |
37.19 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.195 ± 0.062 | 1.533 ± 0.024 |
5.972 ± 0.036 | 6.698 ± 0.047 |
3.952 ± 0.03 | 7.932 ± 0.047 |
1.837 ± 0.02 | 5.335 ± 0.037 |
3.951 ± 0.035 | 9.299 ± 0.061 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.666 ± 0.023 | 3.115 ± 0.032 |
4.302 ± 0.031 | 2.989 ± 0.027 |
5.529 ± 0.067 | 5.922 ± 0.043 |
5.607 ± 0.05 | 8.109 ± 0.046 |
1.097 ± 0.016 | 2.96 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
