
human papillomavirus 164
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Gammapapillomavirus; Gammapapillomavirus 8
Average proteome isoelectric point is 6.02
Get precalculated fractions of proteins
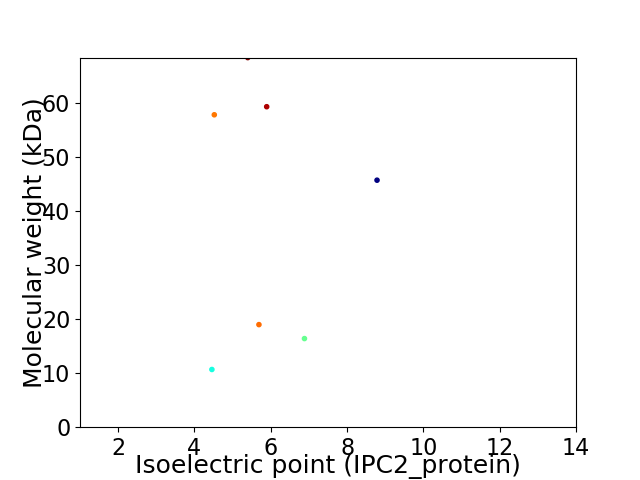
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|K4MN90|K4MN90_9PAPI Major capsid protein L1 OS=human papillomavirus 164 OX=1315261 GN=L1 PE=3 SV=1
MM1 pKa = 7.8IGPQPTKK8 pKa = 10.84EE9 pKa = 4.12NLDD12 pKa = 3.27IDD14 pKa = 4.48LEE16 pKa = 4.31EE17 pKa = 4.86LVLPQNLIAEE27 pKa = 4.67EE28 pKa = 4.24SLSPDD33 pKa = 2.83VDD35 pKa = 3.68PEE37 pKa = 4.1EE38 pKa = 5.19EE39 pKa = 3.94EE40 pKa = 4.51RR41 pKa = 11.84QPFWVDD47 pKa = 3.63TYY49 pKa = 11.39CGTCNASVRR58 pKa = 11.84VSILATASAVCLLRR72 pKa = 11.84VLLQGEE78 pKa = 4.62LCFVCTGCSKK88 pKa = 10.75GRR90 pKa = 11.84LRR92 pKa = 11.84NGRR95 pKa = 11.84SNN97 pKa = 3.19
MM1 pKa = 7.8IGPQPTKK8 pKa = 10.84EE9 pKa = 4.12NLDD12 pKa = 3.27IDD14 pKa = 4.48LEE16 pKa = 4.31EE17 pKa = 4.86LVLPQNLIAEE27 pKa = 4.67EE28 pKa = 4.24SLSPDD33 pKa = 2.83VDD35 pKa = 3.68PEE37 pKa = 4.1EE38 pKa = 5.19EE39 pKa = 3.94EE40 pKa = 4.51RR41 pKa = 11.84QPFWVDD47 pKa = 3.63TYY49 pKa = 11.39CGTCNASVRR58 pKa = 11.84VSILATASAVCLLRR72 pKa = 11.84VLLQGEE78 pKa = 4.62LCFVCTGCSKK88 pKa = 10.75GRR90 pKa = 11.84LRR92 pKa = 11.84NGRR95 pKa = 11.84SNN97 pKa = 3.19
Molecular weight: 10.67 kDa
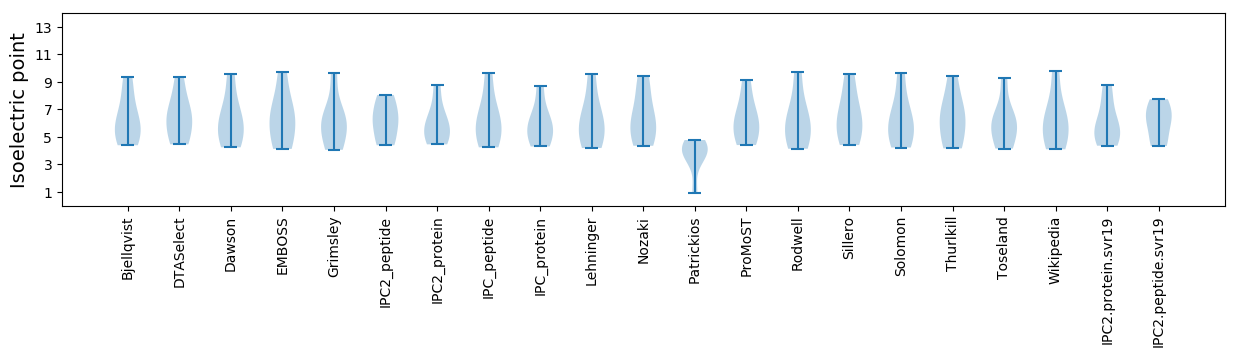
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K4MYW1|K4MYW1_9PAPI Protein E6 OS=human papillomavirus 164 OX=1315261 GN=E6 PE=3 SV=1
MM1 pKa = 7.7VEE3 pKa = 4.04TQEE6 pKa = 4.39TLTQRR11 pKa = 11.84FNALQEE17 pKa = 4.35EE18 pKa = 4.43ILNIIEE24 pKa = 4.81EE25 pKa = 4.61SPTDD29 pKa = 3.72LNSIIKK35 pKa = 9.41YY36 pKa = 7.1WQLTRR41 pKa = 11.84KK42 pKa = 9.82EE43 pKa = 4.33YY44 pKa = 9.19VTLYY48 pKa = 10.12YY49 pKa = 10.54ARR51 pKa = 11.84KK52 pKa = 9.74EE53 pKa = 4.05NITRR57 pKa = 11.84LGLQPVPTLTVSEE70 pKa = 4.64YY71 pKa = 9.63KK72 pKa = 10.64AKK74 pKa = 10.42EE75 pKa = 4.24AIHH78 pKa = 5.68MQLLLTSLSRR88 pKa = 11.84SRR90 pKa = 11.84FASEE94 pKa = 3.23TWTLQDD100 pKa = 4.89CSAEE104 pKa = 4.5LINTQPKK111 pKa = 10.05NCFKK115 pKa = 11.24KK116 pKa = 9.46MGYY119 pKa = 7.46TVDD122 pKa = 2.43VWFDD126 pKa = 3.35HH127 pKa = 7.05DD128 pKa = 4.02RR129 pKa = 11.84SKK131 pKa = 11.56AFPYY135 pKa = 9.93TNWKK139 pKa = 9.34LIYY142 pKa = 9.95YY143 pKa = 9.7QDD145 pKa = 5.93SNDD148 pKa = 3.03QWQKK152 pKa = 9.41TKK154 pKa = 11.51GEE156 pKa = 3.96VDD158 pKa = 3.6EE159 pKa = 5.86NGLFYY164 pKa = 11.44NEE166 pKa = 4.48LNGDD170 pKa = 3.44RR171 pKa = 11.84VYY173 pKa = 10.39FTLFEE178 pKa = 5.13ADD180 pKa = 3.19SAKK183 pKa = 10.89YY184 pKa = 9.72GVSGEE189 pKa = 4.19WTVHH193 pKa = 4.93YY194 pKa = 10.25EE195 pKa = 3.72NTTVVSSSSSNRR207 pKa = 11.84RR208 pKa = 11.84LEE210 pKa = 4.13QPSQTTFDD218 pKa = 3.86EE219 pKa = 5.12PSTSRR224 pKa = 11.84DD225 pKa = 3.24TEE227 pKa = 3.9AQSPKK232 pKa = 9.9RR233 pKa = 11.84RR234 pKa = 11.84RR235 pKa = 11.84QEE237 pKa = 3.73ATSTSSSPSSSTSNLRR253 pKa = 11.84AGRR256 pKa = 11.84RR257 pKa = 11.84GQQQGEE263 pKa = 4.12RR264 pKa = 11.84AAPVPKK270 pKa = 9.89RR271 pKa = 11.84RR272 pKa = 11.84RR273 pKa = 11.84TATSTTQSPEE283 pKa = 3.57RR284 pKa = 11.84FAVPTPSEE292 pKa = 3.84VGTRR296 pKa = 11.84HH297 pKa = 6.69RR298 pKa = 11.84LPPQQGLGRR307 pKa = 11.84LRR309 pKa = 11.84QLQAEE314 pKa = 4.13AWDD317 pKa = 4.45PYY319 pKa = 11.59LLILKK324 pKa = 10.35GSANNLKK331 pKa = 10.18CWRR334 pKa = 11.84NKK336 pKa = 9.63HH337 pKa = 4.94KK338 pKa = 10.56HH339 pKa = 6.01KK340 pKa = 10.86NWFSNSSNVFRR351 pKa = 11.84WLGHH355 pKa = 5.81GCTQSRR361 pKa = 11.84MLLVFDD367 pKa = 4.64TPLQRR372 pKa = 11.84EE373 pKa = 4.11RR374 pKa = 11.84FVMNVRR380 pKa = 11.84LPKK383 pKa = 9.48NTTYY387 pKa = 11.03AYY389 pKa = 10.59GSLDD393 pKa = 3.38SLL395 pKa = 4.41
MM1 pKa = 7.7VEE3 pKa = 4.04TQEE6 pKa = 4.39TLTQRR11 pKa = 11.84FNALQEE17 pKa = 4.35EE18 pKa = 4.43ILNIIEE24 pKa = 4.81EE25 pKa = 4.61SPTDD29 pKa = 3.72LNSIIKK35 pKa = 9.41YY36 pKa = 7.1WQLTRR41 pKa = 11.84KK42 pKa = 9.82EE43 pKa = 4.33YY44 pKa = 9.19VTLYY48 pKa = 10.12YY49 pKa = 10.54ARR51 pKa = 11.84KK52 pKa = 9.74EE53 pKa = 4.05NITRR57 pKa = 11.84LGLQPVPTLTVSEE70 pKa = 4.64YY71 pKa = 9.63KK72 pKa = 10.64AKK74 pKa = 10.42EE75 pKa = 4.24AIHH78 pKa = 5.68MQLLLTSLSRR88 pKa = 11.84SRR90 pKa = 11.84FASEE94 pKa = 3.23TWTLQDD100 pKa = 4.89CSAEE104 pKa = 4.5LINTQPKK111 pKa = 10.05NCFKK115 pKa = 11.24KK116 pKa = 9.46MGYY119 pKa = 7.46TVDD122 pKa = 2.43VWFDD126 pKa = 3.35HH127 pKa = 7.05DD128 pKa = 4.02RR129 pKa = 11.84SKK131 pKa = 11.56AFPYY135 pKa = 9.93TNWKK139 pKa = 9.34LIYY142 pKa = 9.95YY143 pKa = 9.7QDD145 pKa = 5.93SNDD148 pKa = 3.03QWQKK152 pKa = 9.41TKK154 pKa = 11.51GEE156 pKa = 3.96VDD158 pKa = 3.6EE159 pKa = 5.86NGLFYY164 pKa = 11.44NEE166 pKa = 4.48LNGDD170 pKa = 3.44RR171 pKa = 11.84VYY173 pKa = 10.39FTLFEE178 pKa = 5.13ADD180 pKa = 3.19SAKK183 pKa = 10.89YY184 pKa = 9.72GVSGEE189 pKa = 4.19WTVHH193 pKa = 4.93YY194 pKa = 10.25EE195 pKa = 3.72NTTVVSSSSSNRR207 pKa = 11.84RR208 pKa = 11.84LEE210 pKa = 4.13QPSQTTFDD218 pKa = 3.86EE219 pKa = 5.12PSTSRR224 pKa = 11.84DD225 pKa = 3.24TEE227 pKa = 3.9AQSPKK232 pKa = 9.9RR233 pKa = 11.84RR234 pKa = 11.84RR235 pKa = 11.84QEE237 pKa = 3.73ATSTSSSPSSSTSNLRR253 pKa = 11.84AGRR256 pKa = 11.84RR257 pKa = 11.84GQQQGEE263 pKa = 4.12RR264 pKa = 11.84AAPVPKK270 pKa = 9.89RR271 pKa = 11.84RR272 pKa = 11.84RR273 pKa = 11.84TATSTTQSPEE283 pKa = 3.57RR284 pKa = 11.84FAVPTPSEE292 pKa = 3.84VGTRR296 pKa = 11.84HH297 pKa = 6.69RR298 pKa = 11.84LPPQQGLGRR307 pKa = 11.84LRR309 pKa = 11.84QLQAEE314 pKa = 4.13AWDD317 pKa = 4.45PYY319 pKa = 11.59LLILKK324 pKa = 10.35GSANNLKK331 pKa = 10.18CWRR334 pKa = 11.84NKK336 pKa = 9.63HH337 pKa = 4.94KK338 pKa = 10.56HH339 pKa = 6.01KK340 pKa = 10.86NWFSNSSNVFRR351 pKa = 11.84WLGHH355 pKa = 5.81GCTQSRR361 pKa = 11.84MLLVFDD367 pKa = 4.64TPLQRR372 pKa = 11.84EE373 pKa = 4.11RR374 pKa = 11.84FVMNVRR380 pKa = 11.84LPKK383 pKa = 9.48NTTYY387 pKa = 11.03AYY389 pKa = 10.59GSLDD393 pKa = 3.38SLL395 pKa = 4.41
Molecular weight: 45.66 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2443 |
97 |
600 |
349.0 |
39.57 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.69 ± 0.51 | 2.415 ± 0.861 |
6.427 ± 0.542 | 6.427 ± 0.778 |
4.748 ± 0.533 | 4.953 ± 0.607 |
1.76 ± 0.262 | 5.158 ± 0.942 |
5.158 ± 0.756 | 9.742 ± 0.541 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.433 ± 0.289 | 5.239 ± 0.376 |
6.181 ± 1.141 | 4.544 ± 0.7 |
5.813 ± 0.67 | 7.327 ± 0.594 |
6.222 ± 0.927 | 6.099 ± 0.63 |
1.31 ± 0.403 | 3.357 ± 0.228 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
