
Torque teno mini virus ALH8
Taxonomy: Viruses; Anelloviridae; Betatorquevirus; Torque teno mini virus 16
Average proteome isoelectric point is 6.71
Get precalculated fractions of proteins
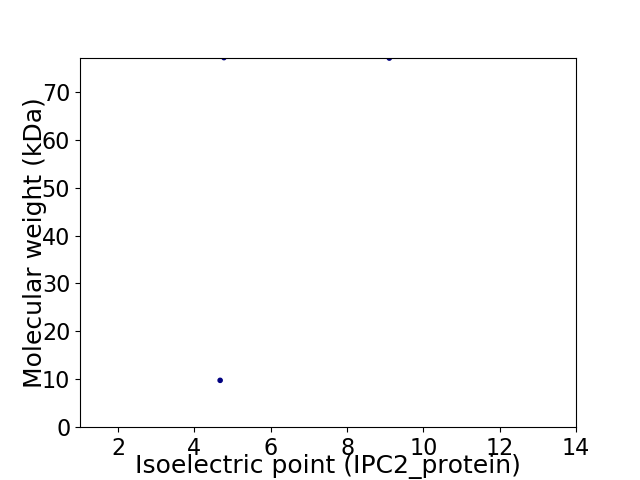
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A076V5S0|A0A076V5S0_9VIRU Uncharacterized protein OS=Torque teno mini virus ALH8 OX=1535291 GN=ALH8_gp1 PE=4 SV=1
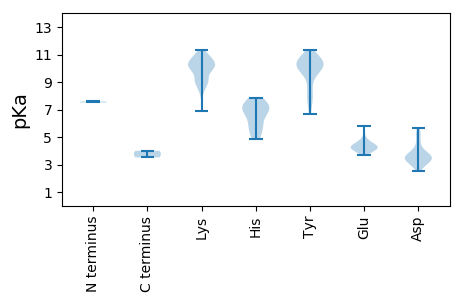
MM1 pKa = 7.51HH2 pKa = 7.85KK3 pKa = 9.62LTRR6 pKa = 11.84AQQIKK11 pKa = 8.2FTNCTVGIHH20 pKa = 7.41DD21 pKa = 5.23CLCDD25 pKa = 3.72CDD27 pKa = 5.68KK28 pKa = 10.84PALHH32 pKa = 7.11AATILLKK39 pKa = 10.66QLGPEE44 pKa = 4.39LLQEE48 pKa = 4.61EE49 pKa = 4.67KK50 pKa = 11.09DD51 pKa = 3.47HH52 pKa = 7.37LKK54 pKa = 10.6RR55 pKa = 11.84CLGTTTGPVEE65 pKa = 4.28EE66 pKa = 4.85DD67 pKa = 3.43CGVTGEE73 pKa = 4.51DD74 pKa = 3.85LEE76 pKa = 4.59KK77 pKa = 11.09LFAEE81 pKa = 4.83DD82 pKa = 3.41TGEE85 pKa = 4.34DD86 pKa = 3.32QDD88 pKa = 4.06GG89 pKa = 3.52
MM1 pKa = 7.51HH2 pKa = 7.85KK3 pKa = 9.62LTRR6 pKa = 11.84AQQIKK11 pKa = 8.2FTNCTVGIHH20 pKa = 7.41DD21 pKa = 5.23CLCDD25 pKa = 3.72CDD27 pKa = 5.68KK28 pKa = 10.84PALHH32 pKa = 7.11AATILLKK39 pKa = 10.66QLGPEE44 pKa = 4.39LLQEE48 pKa = 4.61EE49 pKa = 4.67KK50 pKa = 11.09DD51 pKa = 3.47HH52 pKa = 7.37LKK54 pKa = 10.6RR55 pKa = 11.84CLGTTTGPVEE65 pKa = 4.28EE66 pKa = 4.85DD67 pKa = 3.43CGVTGEE73 pKa = 4.51DD74 pKa = 3.85LEE76 pKa = 4.59KK77 pKa = 11.09LFAEE81 pKa = 4.83DD82 pKa = 3.41TGEE85 pKa = 4.34DD86 pKa = 3.32QDD88 pKa = 4.06GG89 pKa = 3.52
Molecular weight: 9.78 kDa
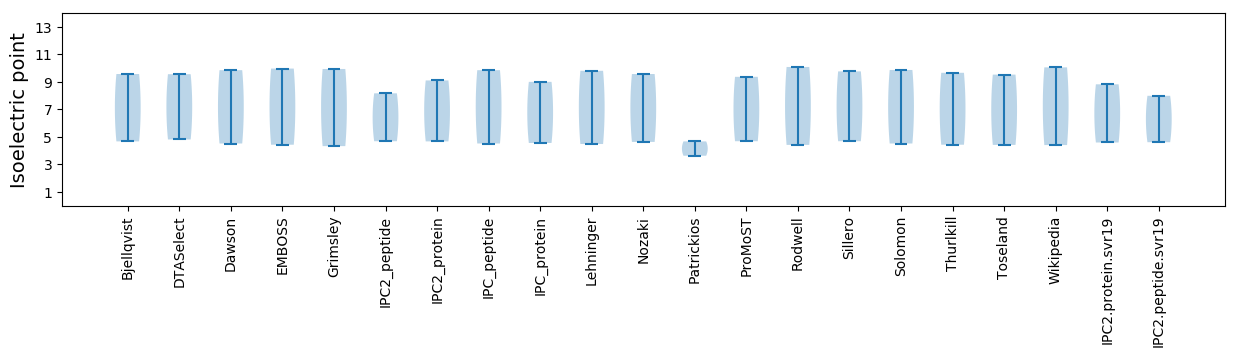
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A076V5S0|A0A076V5S0_9VIRU Uncharacterized protein OS=Torque teno mini virus ALH8 OX=1535291 GN=ALH8_gp1 PE=4 SV=1
MM1 pKa = 7.58PWYY4 pKa = 10.17NYY6 pKa = 7.45WPRR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84LRR14 pKa = 11.84RR15 pKa = 11.84YY16 pKa = 6.68WRR18 pKa = 11.84RR19 pKa = 11.84PRR21 pKa = 11.84KK22 pKa = 6.91TLRR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84YY28 pKa = 7.5WRR30 pKa = 11.84RR31 pKa = 11.84PRR33 pKa = 11.84WVRR36 pKa = 11.84RR37 pKa = 11.84PYY39 pKa = 10.14KK40 pKa = 10.1RR41 pKa = 11.84KK42 pKa = 9.28LRR44 pKa = 11.84KK45 pKa = 8.97IVLTQFQPKK54 pKa = 8.98TIRR57 pKa = 11.84KK58 pKa = 8.78SKK60 pKa = 10.38IKK62 pKa = 10.81GPICLFQTTNKK73 pKa = 10.22RR74 pKa = 11.84LVNNFDD80 pKa = 3.38MYY82 pKa = 9.89EE83 pKa = 3.94TSEE86 pKa = 4.36VPEE89 pKa = 4.13HH90 pKa = 5.94QPGGGGWGIKK100 pKa = 10.13VFTLEE105 pKa = 4.03GLYY108 pKa = 10.29SEE110 pKa = 5.82HH111 pKa = 7.54IYY113 pKa = 10.75GRR115 pKa = 11.84NIWTVTNQDD124 pKa = 3.73LPLVRR129 pKa = 11.84YY130 pKa = 8.33LGCSIRR136 pKa = 11.84FYY138 pKa = 10.53QSEE141 pKa = 4.35YY142 pKa = 7.9TDD144 pKa = 3.79YY145 pKa = 11.36VATYY149 pKa = 10.69SNQLPLQSSLGMYY162 pKa = 10.59NAMQPNIHH170 pKa = 6.84LLLKK174 pKa = 10.83NKK176 pKa = 8.46ITMPGLKK183 pKa = 8.92TYY185 pKa = 9.67KK186 pKa = 10.25RR187 pKa = 11.84KK188 pKa = 10.12VPYY191 pKa = 9.83KK192 pKa = 10.09KK193 pKa = 10.7VFVPPPTQLEE203 pKa = 4.38NKK205 pKa = 9.11WYY207 pKa = 9.57FQQNMAKK214 pKa = 9.01TPLLMTRR221 pKa = 11.84VTACSLNKK229 pKa = 10.04FYY231 pKa = 10.57IDD233 pKa = 3.79PDD235 pKa = 4.3HH236 pKa = 7.23INTNLTITSLNVSLFSNRR254 pKa = 11.84QFQDD258 pKa = 2.76ATDD261 pKa = 3.67YY262 pKa = 11.13HH263 pKa = 6.95PKK265 pKa = 9.3TVGTINYY272 pKa = 8.65YY273 pKa = 10.6LYY275 pKa = 9.9STTQNPPYY283 pKa = 10.36NGPLKK288 pKa = 10.77LKK290 pKa = 9.85MLIPLTDD297 pKa = 3.22TMIYY301 pKa = 10.18KK302 pKa = 10.4PGINYY307 pKa = 9.92EE308 pKa = 3.97EE309 pKa = 4.38YY310 pKa = 10.59KK311 pKa = 10.86RR312 pKa = 11.84NNTTKK317 pKa = 10.27SWTDD321 pKa = 2.52WKK323 pKa = 9.94TEE325 pKa = 3.7YY326 pKa = 10.22TIYY329 pKa = 10.69AGNPFHH335 pKa = 6.87TNYY338 pKa = 10.39LSVGEE343 pKa = 4.03PVFLIGKK350 pKa = 9.15KK351 pKa = 10.15PSDD354 pKa = 3.52LFSSEE359 pKa = 4.15EE360 pKa = 4.55GEE362 pKa = 4.25TSDD365 pKa = 3.57YY366 pKa = 7.91TTAEE370 pKa = 3.83LTKK373 pKa = 9.73TIRR376 pKa = 11.84YY377 pKa = 8.66NPYY380 pKa = 9.99NDD382 pKa = 3.44QGQTNICYY390 pKa = 10.23FKK392 pKa = 11.35ANFKK396 pKa = 11.0SEE398 pKa = 4.4TSWQPPDD405 pKa = 4.59NPDD408 pKa = 3.26LTNEE412 pKa = 4.21NLPFWLLLWGFSDD425 pKa = 3.17WHH427 pKa = 6.21KK428 pKa = 10.71KK429 pKa = 8.36IKK431 pKa = 9.96KK432 pKa = 8.83HH433 pKa = 5.17LHH435 pKa = 5.83IDD437 pKa = 3.07SSYY440 pKa = 9.51ILVMRR445 pKa = 11.84HH446 pKa = 5.73IPLALNTEE454 pKa = 4.63YY455 pKa = 10.25IVPLSDD461 pKa = 4.88SFLQGKK467 pKa = 9.31SPFSPEE473 pKa = 3.92EE474 pKa = 3.71QPIGADD480 pKa = 2.84RR481 pKa = 11.84TTWHH485 pKa = 6.84PQFQHH490 pKa = 4.83QAEE493 pKa = 4.96AINTICSAGPGTTKK507 pKa = 10.32IPDD510 pKa = 3.59NYY512 pKa = 10.6SVQGLMKK519 pKa = 10.45YY520 pKa = 10.3SFYY523 pKa = 10.72FKK525 pKa = 9.98WGGSPPPMSTIEE537 pKa = 5.36DD538 pKa = 5.32PIQQPTYY545 pKa = 9.7PVPHH549 pKa = 6.77NFNLTTSLQNPEE561 pKa = 4.17TDD563 pKa = 3.31PASILWSFDD572 pKa = 3.14QRR574 pKa = 11.84RR575 pKa = 11.84HH576 pKa = 5.33SLTKK580 pKa = 10.17KK581 pKa = 10.43AIEE584 pKa = 4.19RR585 pKa = 11.84LQKK588 pKa = 9.94DD589 pKa = 3.48TGIKK593 pKa = 9.41TITVTGGSHH602 pKa = 7.55FDD604 pKa = 3.54PAPTISQEE612 pKa = 4.17EE613 pKa = 4.26TSEE616 pKa = 4.06EE617 pKa = 3.98SSEE620 pKa = 4.18EE621 pKa = 3.95EE622 pKa = 4.39KK623 pKa = 10.17EE624 pKa = 4.28TSLLEE629 pKa = 4.58KK630 pKa = 10.39LQQQQRR636 pKa = 11.84QQRR639 pKa = 11.84RR640 pKa = 11.84LKK642 pKa = 10.36LRR644 pKa = 11.84IMKK647 pKa = 8.95TLLKK651 pKa = 10.08IQQLEE656 pKa = 4.0
MM1 pKa = 7.58PWYY4 pKa = 10.17NYY6 pKa = 7.45WPRR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84LRR14 pKa = 11.84RR15 pKa = 11.84YY16 pKa = 6.68WRR18 pKa = 11.84RR19 pKa = 11.84PRR21 pKa = 11.84KK22 pKa = 6.91TLRR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84YY28 pKa = 7.5WRR30 pKa = 11.84RR31 pKa = 11.84PRR33 pKa = 11.84WVRR36 pKa = 11.84RR37 pKa = 11.84PYY39 pKa = 10.14KK40 pKa = 10.1RR41 pKa = 11.84KK42 pKa = 9.28LRR44 pKa = 11.84KK45 pKa = 8.97IVLTQFQPKK54 pKa = 8.98TIRR57 pKa = 11.84KK58 pKa = 8.78SKK60 pKa = 10.38IKK62 pKa = 10.81GPICLFQTTNKK73 pKa = 10.22RR74 pKa = 11.84LVNNFDD80 pKa = 3.38MYY82 pKa = 9.89EE83 pKa = 3.94TSEE86 pKa = 4.36VPEE89 pKa = 4.13HH90 pKa = 5.94QPGGGGWGIKK100 pKa = 10.13VFTLEE105 pKa = 4.03GLYY108 pKa = 10.29SEE110 pKa = 5.82HH111 pKa = 7.54IYY113 pKa = 10.75GRR115 pKa = 11.84NIWTVTNQDD124 pKa = 3.73LPLVRR129 pKa = 11.84YY130 pKa = 8.33LGCSIRR136 pKa = 11.84FYY138 pKa = 10.53QSEE141 pKa = 4.35YY142 pKa = 7.9TDD144 pKa = 3.79YY145 pKa = 11.36VATYY149 pKa = 10.69SNQLPLQSSLGMYY162 pKa = 10.59NAMQPNIHH170 pKa = 6.84LLLKK174 pKa = 10.83NKK176 pKa = 8.46ITMPGLKK183 pKa = 8.92TYY185 pKa = 9.67KK186 pKa = 10.25RR187 pKa = 11.84KK188 pKa = 10.12VPYY191 pKa = 9.83KK192 pKa = 10.09KK193 pKa = 10.7VFVPPPTQLEE203 pKa = 4.38NKK205 pKa = 9.11WYY207 pKa = 9.57FQQNMAKK214 pKa = 9.01TPLLMTRR221 pKa = 11.84VTACSLNKK229 pKa = 10.04FYY231 pKa = 10.57IDD233 pKa = 3.79PDD235 pKa = 4.3HH236 pKa = 7.23INTNLTITSLNVSLFSNRR254 pKa = 11.84QFQDD258 pKa = 2.76ATDD261 pKa = 3.67YY262 pKa = 11.13HH263 pKa = 6.95PKK265 pKa = 9.3TVGTINYY272 pKa = 8.65YY273 pKa = 10.6LYY275 pKa = 9.9STTQNPPYY283 pKa = 10.36NGPLKK288 pKa = 10.77LKK290 pKa = 9.85MLIPLTDD297 pKa = 3.22TMIYY301 pKa = 10.18KK302 pKa = 10.4PGINYY307 pKa = 9.92EE308 pKa = 3.97EE309 pKa = 4.38YY310 pKa = 10.59KK311 pKa = 10.86RR312 pKa = 11.84NNTTKK317 pKa = 10.27SWTDD321 pKa = 2.52WKK323 pKa = 9.94TEE325 pKa = 3.7YY326 pKa = 10.22TIYY329 pKa = 10.69AGNPFHH335 pKa = 6.87TNYY338 pKa = 10.39LSVGEE343 pKa = 4.03PVFLIGKK350 pKa = 9.15KK351 pKa = 10.15PSDD354 pKa = 3.52LFSSEE359 pKa = 4.15EE360 pKa = 4.55GEE362 pKa = 4.25TSDD365 pKa = 3.57YY366 pKa = 7.91TTAEE370 pKa = 3.83LTKK373 pKa = 9.73TIRR376 pKa = 11.84YY377 pKa = 8.66NPYY380 pKa = 9.99NDD382 pKa = 3.44QGQTNICYY390 pKa = 10.23FKK392 pKa = 11.35ANFKK396 pKa = 11.0SEE398 pKa = 4.4TSWQPPDD405 pKa = 4.59NPDD408 pKa = 3.26LTNEE412 pKa = 4.21NLPFWLLLWGFSDD425 pKa = 3.17WHH427 pKa = 6.21KK428 pKa = 10.71KK429 pKa = 8.36IKK431 pKa = 9.96KK432 pKa = 8.83HH433 pKa = 5.17LHH435 pKa = 5.83IDD437 pKa = 3.07SSYY440 pKa = 9.51ILVMRR445 pKa = 11.84HH446 pKa = 5.73IPLALNTEE454 pKa = 4.63YY455 pKa = 10.25IVPLSDD461 pKa = 4.88SFLQGKK467 pKa = 9.31SPFSPEE473 pKa = 3.92EE474 pKa = 3.71QPIGADD480 pKa = 2.84RR481 pKa = 11.84TTWHH485 pKa = 6.84PQFQHH490 pKa = 4.83QAEE493 pKa = 4.96AINTICSAGPGTTKK507 pKa = 10.32IPDD510 pKa = 3.59NYY512 pKa = 10.6SVQGLMKK519 pKa = 10.45YY520 pKa = 10.3SFYY523 pKa = 10.72FKK525 pKa = 9.98WGGSPPPMSTIEE537 pKa = 5.36DD538 pKa = 5.32PIQQPTYY545 pKa = 9.7PVPHH549 pKa = 6.77NFNLTTSLQNPEE561 pKa = 4.17TDD563 pKa = 3.31PASILWSFDD572 pKa = 3.14QRR574 pKa = 11.84RR575 pKa = 11.84HH576 pKa = 5.33SLTKK580 pKa = 10.17KK581 pKa = 10.43AIEE584 pKa = 4.19RR585 pKa = 11.84LQKK588 pKa = 9.94DD589 pKa = 3.48TGIKK593 pKa = 9.41TITVTGGSHH602 pKa = 7.55FDD604 pKa = 3.54PAPTISQEE612 pKa = 4.17EE613 pKa = 4.26TSEE616 pKa = 4.06EE617 pKa = 3.98SSEE620 pKa = 4.18EE621 pKa = 3.95EE622 pKa = 4.39KK623 pKa = 10.17EE624 pKa = 4.28TSLLEE629 pKa = 4.58KK630 pKa = 10.39LQQQQRR636 pKa = 11.84QQRR639 pKa = 11.84RR640 pKa = 11.84LKK642 pKa = 10.36LRR644 pKa = 11.84IMKK647 pKa = 8.95TLLKK651 pKa = 10.08IQQLEE656 pKa = 4.0
Molecular weight: 77.06 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
745 |
89 |
656 |
372.5 |
43.42 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
2.819 ± 1.317 | 1.477 ± 2.477 |
4.43 ± 2.673 | 5.772 ± 2.042 |
3.624 ± 0.648 | 5.101 ± 1.829 |
2.55 ± 0.914 | 5.638 ± 1.066 |
7.517 ± 0.164 | 9.53 ± 1.859 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.879 ± 0.355 | 5.235 ± 1.934 |
7.383 ± 1.887 | 5.772 ± 0.072 |
5.369 ± 1.469 | 5.772 ± 2.715 |
9.396 ± 0.337 | 3.221 ± 0.07 |
2.282 ± 1.073 | 5.235 ± 2.463 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
