
Bat circovirus ZS/China/2011
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; Cyclovirus; Dragonfly associated cyclovirus 4
Average proteome isoelectric point is 7.57
Get precalculated fractions of proteins
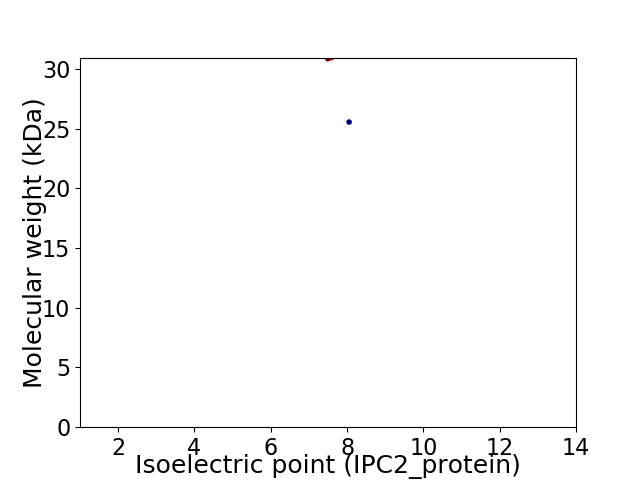
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|G1D7G1|G1D7G1_9CIRC Putative capsid protein OS=Bat circovirus ZS/China/2011 OX=1072162 PE=4 SV=1
MM1 pKa = 7.84PCLQARR7 pKa = 11.84YY8 pKa = 8.35WLLTIPYY15 pKa = 9.12EE16 pKa = 4.19HH17 pKa = 6.85FTPYY21 pKa = 10.66LPPNCAYY28 pKa = 9.81IKK30 pKa = 10.65GQLEE34 pKa = 4.11QGSNTSYY41 pKa = 10.88LHH43 pKa = 5.57WQLVVYY49 pKa = 8.08FSQKK53 pKa = 10.32KK54 pKa = 8.36SLNYY58 pKa = 10.01VKK60 pKa = 10.7LIFGDD65 pKa = 5.23GIHH68 pKa = 6.8CEE70 pKa = 3.95PSKK73 pKa = 11.22SKK75 pKa = 10.7AAEE78 pKa = 3.93EE79 pKa = 4.42YY80 pKa = 9.67VWKK83 pKa = 9.97EE84 pKa = 3.73DD85 pKa = 3.29TSIPGTQFEE94 pKa = 4.78LGKK97 pKa = 10.62NSIKK101 pKa = 10.19RR102 pKa = 11.84DD103 pKa = 3.54STKK106 pKa = 11.07DD107 pKa = 2.47WDD109 pKa = 5.15LIVKK113 pKa = 9.16HH114 pKa = 6.14ARR116 pKa = 11.84EE117 pKa = 4.3GDD119 pKa = 3.63FASIPGDD126 pKa = 3.43VLVRR130 pKa = 11.84CYY132 pKa = 11.49GNLKK136 pKa = 9.9KK137 pKa = 10.4IRR139 pKa = 11.84VDD141 pKa = 3.35SLQPEE146 pKa = 4.59SIVRR150 pKa = 11.84EE151 pKa = 3.69IYY153 pKa = 10.28VYY155 pKa = 10.55YY156 pKa = 10.81GRR158 pKa = 11.84TGAGKK163 pKa = 9.71SRR165 pKa = 11.84RR166 pKa = 11.84AWEE169 pKa = 3.97EE170 pKa = 3.77ATLSAYY176 pKa = 9.81PKK178 pKa = 10.66DD179 pKa = 3.98PNTKK183 pKa = 9.62FWDD186 pKa = 4.78GYY188 pKa = 10.96AGQEE192 pKa = 4.02CVVIDD197 pKa = 4.08EE198 pKa = 4.38FRR200 pKa = 11.84GAISISHH207 pKa = 7.22LLRR210 pKa = 11.84WLDD213 pKa = 3.65RR214 pKa = 11.84YY215 pKa = 9.87PVIVEE220 pKa = 4.16IKK222 pKa = 10.51GSSCVFKK229 pKa = 10.9AKK231 pKa = 10.39KK232 pKa = 9.58IWITSNLSPDD242 pKa = 3.25DD243 pKa = 4.05WYY245 pKa = 10.61PDD247 pKa = 3.23LDD249 pKa = 4.92AEE251 pKa = 4.68TKK253 pKa = 9.62SALRR257 pKa = 11.84RR258 pKa = 11.84RR259 pKa = 11.84FTEE262 pKa = 4.13VVHH265 pKa = 6.78FNN267 pKa = 3.4
MM1 pKa = 7.84PCLQARR7 pKa = 11.84YY8 pKa = 8.35WLLTIPYY15 pKa = 9.12EE16 pKa = 4.19HH17 pKa = 6.85FTPYY21 pKa = 10.66LPPNCAYY28 pKa = 9.81IKK30 pKa = 10.65GQLEE34 pKa = 4.11QGSNTSYY41 pKa = 10.88LHH43 pKa = 5.57WQLVVYY49 pKa = 8.08FSQKK53 pKa = 10.32KK54 pKa = 8.36SLNYY58 pKa = 10.01VKK60 pKa = 10.7LIFGDD65 pKa = 5.23GIHH68 pKa = 6.8CEE70 pKa = 3.95PSKK73 pKa = 11.22SKK75 pKa = 10.7AAEE78 pKa = 3.93EE79 pKa = 4.42YY80 pKa = 9.67VWKK83 pKa = 9.97EE84 pKa = 3.73DD85 pKa = 3.29TSIPGTQFEE94 pKa = 4.78LGKK97 pKa = 10.62NSIKK101 pKa = 10.19RR102 pKa = 11.84DD103 pKa = 3.54STKK106 pKa = 11.07DD107 pKa = 2.47WDD109 pKa = 5.15LIVKK113 pKa = 9.16HH114 pKa = 6.14ARR116 pKa = 11.84EE117 pKa = 4.3GDD119 pKa = 3.63FASIPGDD126 pKa = 3.43VLVRR130 pKa = 11.84CYY132 pKa = 11.49GNLKK136 pKa = 9.9KK137 pKa = 10.4IRR139 pKa = 11.84VDD141 pKa = 3.35SLQPEE146 pKa = 4.59SIVRR150 pKa = 11.84EE151 pKa = 3.69IYY153 pKa = 10.28VYY155 pKa = 10.55YY156 pKa = 10.81GRR158 pKa = 11.84TGAGKK163 pKa = 9.71SRR165 pKa = 11.84RR166 pKa = 11.84AWEE169 pKa = 3.97EE170 pKa = 3.77ATLSAYY176 pKa = 9.81PKK178 pKa = 10.66DD179 pKa = 3.98PNTKK183 pKa = 9.62FWDD186 pKa = 4.78GYY188 pKa = 10.96AGQEE192 pKa = 4.02CVVIDD197 pKa = 4.08EE198 pKa = 4.38FRR200 pKa = 11.84GAISISHH207 pKa = 7.22LLRR210 pKa = 11.84WLDD213 pKa = 3.65RR214 pKa = 11.84YY215 pKa = 9.87PVIVEE220 pKa = 4.16IKK222 pKa = 10.51GSSCVFKK229 pKa = 10.9AKK231 pKa = 10.39KK232 pKa = 9.58IWITSNLSPDD242 pKa = 3.25DD243 pKa = 4.05WYY245 pKa = 10.61PDD247 pKa = 3.23LDD249 pKa = 4.92AEE251 pKa = 4.68TKK253 pKa = 9.62SALRR257 pKa = 11.84RR258 pKa = 11.84RR259 pKa = 11.84FTEE262 pKa = 4.13VVHH265 pKa = 6.78FNN267 pKa = 3.4
Molecular weight: 30.84 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G1D7G1|G1D7G1_9CIRC Putative capsid protein OS=Bat circovirus ZS/China/2011 OX=1072162 PE=4 SV=1
MM1 pKa = 6.47TQVIKK6 pKa = 11.17GMMALTLYY14 pKa = 10.38GINGTNDD21 pKa = 2.78GNTVQGCRR29 pKa = 11.84DD30 pKa = 2.97IYY32 pKa = 10.78RR33 pKa = 11.84IFKK36 pKa = 10.32NDD38 pKa = 3.71PDD40 pKa = 3.92VQQTGVAPGNEE51 pKa = 4.3PYY53 pKa = 10.31NGKK56 pKa = 8.88LQFGSGVLDD65 pKa = 3.23VTIRR69 pKa = 11.84NLSDD73 pKa = 3.67SIEE76 pKa = 4.44AEE78 pKa = 3.74VDD80 pKa = 3.11IYY82 pKa = 10.94TGWFRR87 pKa = 11.84KK88 pKa = 9.86SQDD91 pKa = 2.94NTAVLGSRR99 pKa = 11.84NPVTDD104 pKa = 4.33FSNPSAITPIAPGNTGTQIYY124 pKa = 10.33EE125 pKa = 3.95RR126 pKa = 11.84GTTLFDD132 pKa = 3.25GTTALSKK139 pKa = 9.88TGFHH143 pKa = 6.47IHH145 pKa = 7.04RR146 pKa = 11.84KK147 pKa = 6.68TKK149 pKa = 10.57SILAPTQSMFLQHH162 pKa = 7.26RR163 pKa = 11.84DD164 pKa = 3.8PKK166 pKa = 11.12NHH168 pKa = 6.34MIDD171 pKa = 3.14WVRR174 pKa = 11.84DD175 pKa = 3.62GVVGYY180 pKa = 10.23AKK182 pKa = 10.54KK183 pKa = 10.63GLTYY187 pKa = 11.13GMIIVWKK194 pKa = 9.06PSVLASSEE202 pKa = 4.43AIVPLAVGTTRR213 pKa = 11.84KK214 pKa = 8.13YY215 pKa = 8.63TYY217 pKa = 11.12AVVEE221 pKa = 4.44DD222 pKa = 3.91NLDD225 pKa = 3.77RR226 pKa = 11.84VCKK229 pKa = 10.5NPPLL233 pKa = 4.4
MM1 pKa = 6.47TQVIKK6 pKa = 11.17GMMALTLYY14 pKa = 10.38GINGTNDD21 pKa = 2.78GNTVQGCRR29 pKa = 11.84DD30 pKa = 2.97IYY32 pKa = 10.78RR33 pKa = 11.84IFKK36 pKa = 10.32NDD38 pKa = 3.71PDD40 pKa = 3.92VQQTGVAPGNEE51 pKa = 4.3PYY53 pKa = 10.31NGKK56 pKa = 8.88LQFGSGVLDD65 pKa = 3.23VTIRR69 pKa = 11.84NLSDD73 pKa = 3.67SIEE76 pKa = 4.44AEE78 pKa = 3.74VDD80 pKa = 3.11IYY82 pKa = 10.94TGWFRR87 pKa = 11.84KK88 pKa = 9.86SQDD91 pKa = 2.94NTAVLGSRR99 pKa = 11.84NPVTDD104 pKa = 4.33FSNPSAITPIAPGNTGTQIYY124 pKa = 10.33EE125 pKa = 3.95RR126 pKa = 11.84GTTLFDD132 pKa = 3.25GTTALSKK139 pKa = 9.88TGFHH143 pKa = 6.47IHH145 pKa = 7.04RR146 pKa = 11.84KK147 pKa = 6.68TKK149 pKa = 10.57SILAPTQSMFLQHH162 pKa = 7.26RR163 pKa = 11.84DD164 pKa = 3.8PKK166 pKa = 11.12NHH168 pKa = 6.34MIDD171 pKa = 3.14WVRR174 pKa = 11.84DD175 pKa = 3.62GVVGYY180 pKa = 10.23AKK182 pKa = 10.54KK183 pKa = 10.63GLTYY187 pKa = 11.13GMIIVWKK194 pKa = 9.06PSVLASSEE202 pKa = 4.43AIVPLAVGTTRR213 pKa = 11.84KK214 pKa = 8.13YY215 pKa = 8.63TYY217 pKa = 11.12AVVEE221 pKa = 4.44DD222 pKa = 3.91NLDD225 pKa = 3.77RR226 pKa = 11.84VCKK229 pKa = 10.5NPPLL233 pKa = 4.4
Molecular weight: 25.54 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
500 |
233 |
267 |
250.0 |
28.19 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.6 ± 0.013 | 1.6 ± 0.453 |
6.2 ± 0.145 | 4.6 ± 1.236 |
3.4 ± 0.241 | 7.6 ± 1.124 |
2.0 ± 0.173 | 6.8 ± 0.041 |
6.8 ± 0.745 | 7.2 ± 0.465 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.4 ± 0.717 | 4.4 ± 0.982 |
5.4 ± 0.109 | 3.4 ± 0.282 |
5.2 ± 0.292 | 6.8 ± 0.745 |
7.0 ± 1.752 | 7.2 ± 0.582 |
2.4 ± 0.679 | 5.0 ± 0.694 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
