
Maritimibacter alkaliphilus HTCC2654
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Roseobacteraceae; Maritimibacter; Maritimibacter alkaliphilus
Average proteome isoelectric point is 6.09
Get precalculated fractions of proteins
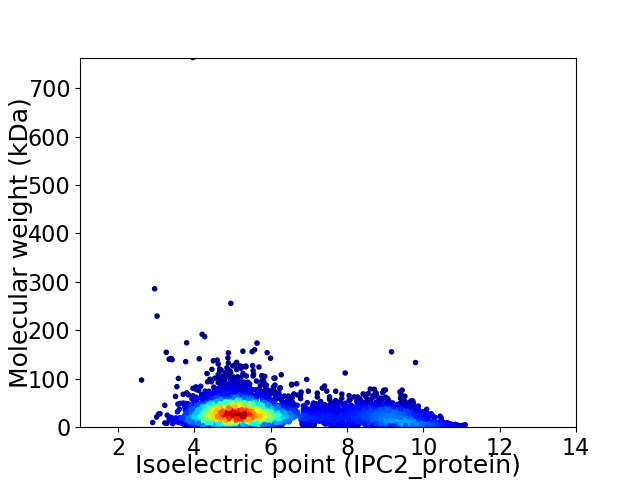
Virtual 2D-PAGE plot for 4692 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A3VIA4|A3VIA4_9RHOB Uncharacterized protein OS=Maritimibacter alkaliphilus HTCC2654 OX=314271 GN=RB2654_01335 PE=4 SV=1
MM1 pKa = 7.28ATMNSGLGGPAGYY14 pKa = 10.35GEE16 pKa = 4.82NVFSSTSKK24 pKa = 7.72TTGNNDD30 pKa = 3.45DD31 pKa = 3.57GSVYY35 pKa = 10.74VDD37 pKa = 3.52TSSVFGDD44 pKa = 4.28GIDD47 pKa = 4.04FFGTTYY53 pKa = 10.75SGLYY57 pKa = 9.7VNSNGNITFGAANTAYY73 pKa = 8.51QTSNLAGTSSRR84 pKa = 11.84MIAPFWGDD92 pKa = 3.18VNINSGGEE100 pKa = 4.09IYY102 pKa = 10.12WDD104 pKa = 3.44LDD106 pKa = 3.37AAAGTVTITWDD117 pKa = 3.22GVAPYY122 pKa = 9.69NGSGANSFQIVLTDD136 pKa = 3.2IGGGDD141 pKa = 3.52FTVEE145 pKa = 5.05FIYY148 pKa = 10.85EE149 pKa = 4.47DD150 pKa = 3.71IQWTTGYY157 pKa = 10.09SQVAQAGITDD167 pKa = 4.1GGANDD172 pKa = 5.44FILPGSGNATTMSNYY187 pKa = 6.77EE188 pKa = 4.13TYY190 pKa = 11.07DD191 pKa = 3.25FGTNDD196 pKa = 3.69PNGTTDD202 pKa = 4.04FFFVNGQPVFGDD214 pKa = 3.78GVVEE218 pKa = 4.43GTSGADD224 pKa = 3.46TIDD227 pKa = 2.98AAYY230 pKa = 10.83YY231 pKa = 10.48GDD233 pKa = 4.63PDD235 pKa = 4.74GDD237 pKa = 4.95LIDD240 pKa = 5.87DD241 pKa = 4.48GDD243 pKa = 4.18GTGPAGNEE251 pKa = 3.53DD252 pKa = 4.88LIYY255 pKa = 10.95GFDD258 pKa = 3.64GDD260 pKa = 5.03DD261 pKa = 4.22NIFAGDD267 pKa = 4.13GDD269 pKa = 3.92DD270 pKa = 4.2TIYY273 pKa = 11.3GGDD276 pKa = 3.48GTDD279 pKa = 3.77NIFGEE284 pKa = 4.96DD285 pKa = 4.02GDD287 pKa = 4.04DD288 pKa = 4.37TIYY291 pKa = 11.25GDD293 pKa = 3.81NGPTGVTEE301 pKa = 4.24DD302 pKa = 4.86LNWSAQGADD311 pKa = 3.51GASIAGGFTQTTGTMDD327 pKa = 3.39VNVSFSADD335 pKa = 2.89GNNAVFEE342 pKa = 4.41VATAEE347 pKa = 4.22TNYY350 pKa = 10.37VAGGEE355 pKa = 4.02PFAPNSSAYY364 pKa = 10.65VYY366 pKa = 11.2GNGDD370 pKa = 3.7GPTGTVTIDD379 pKa = 3.53FAAVPGSGMSDD390 pKa = 3.13EE391 pKa = 4.45VQNVAFRR398 pKa = 11.84LNDD401 pKa = 3.16IDD403 pKa = 5.12SYY405 pKa = 11.48AGNHH409 pKa = 6.18IDD411 pKa = 3.51VLTVNAYY418 pKa = 10.12DD419 pKa = 3.86EE420 pKa = 5.09NGDD423 pKa = 3.86PVAVTLTAAGNDD435 pKa = 3.65SVSGNTVTAGGTLDD449 pKa = 4.22NPQDD453 pKa = 3.66AQGSVLVEE461 pKa = 3.59IDD463 pKa = 4.38GPVAWFEE470 pKa = 4.31VIYY473 pKa = 11.1VNALTGTHH481 pKa = 7.55GINITDD487 pKa = 3.53VSFDD491 pKa = 3.96TIPASGAADD500 pKa = 3.79NIFGGLGDD508 pKa = 3.96DD509 pKa = 4.61TIHH512 pKa = 7.14GEE514 pKa = 4.51AGDD517 pKa = 3.82DD518 pKa = 4.39VIWGDD523 pKa = 3.91EE524 pKa = 4.26GSDD527 pKa = 3.51TLFGGAGDD535 pKa = 3.8DD536 pKa = 4.47TIWAGAGDD544 pKa = 4.67TVTGGAGDD552 pKa = 3.94DD553 pKa = 4.4MIWLDD558 pKa = 3.88AANALGGPGAVIDD571 pKa = 4.94IIGSEE576 pKa = 4.14DD577 pKa = 3.96GEE579 pKa = 4.74TGGDD583 pKa = 3.47TLNFAGLIDD592 pKa = 4.22WGTISYY598 pKa = 9.89TDD600 pKa = 4.03AEE602 pKa = 4.71SGSATLSDD610 pKa = 3.62GTTVTFSNIEE620 pKa = 3.74NLIICFTGGTAIDD633 pKa = 4.41TPSGPCPVEE642 pKa = 3.57HH643 pKa = 6.81LRR645 pKa = 11.84PGDD648 pKa = 3.61LVLTRR653 pKa = 11.84DD654 pKa = 4.33RR655 pKa = 11.84GPQPIRR661 pKa = 11.84WIGRR665 pKa = 11.84RR666 pKa = 11.84TVPGTGTFAPIRR678 pKa = 11.84FEE680 pKa = 4.44RR681 pKa = 11.84GAFGNVAPLMVSPQHH696 pKa = 6.25RR697 pKa = 11.84MVYY700 pKa = 9.77SGSRR704 pKa = 11.84ANLYY708 pKa = 10.41FDD710 pKa = 3.84TPEE713 pKa = 3.73VMVPAKK719 pKa = 10.5HH720 pKa = 6.64LVDD723 pKa = 3.36QRR725 pKa = 11.84LIRR728 pKa = 11.84QVEE731 pKa = 4.63VPTVTYY737 pKa = 8.48FHH739 pKa = 7.12IMLDD743 pKa = 3.2RR744 pKa = 11.84HH745 pKa = 5.22EE746 pKa = 4.46VLWGNGAPSEE756 pKa = 4.4SFHH759 pKa = 6.78PGALGLDD766 pKa = 3.48ALAAEE771 pKa = 4.58ARR773 pKa = 11.84EE774 pKa = 4.05EE775 pKa = 4.13LLTLFPDD782 pKa = 3.97LRR784 pKa = 11.84ADD786 pKa = 3.45PGVYY790 pKa = 10.14GKK792 pKa = 7.46TARR795 pKa = 11.84RR796 pKa = 11.84VLKK799 pKa = 10.5RR800 pKa = 11.84FEE802 pKa = 4.18ARR804 pKa = 11.84LLAAA808 pKa = 5.49
MM1 pKa = 7.28ATMNSGLGGPAGYY14 pKa = 10.35GEE16 pKa = 4.82NVFSSTSKK24 pKa = 7.72TTGNNDD30 pKa = 3.45DD31 pKa = 3.57GSVYY35 pKa = 10.74VDD37 pKa = 3.52TSSVFGDD44 pKa = 4.28GIDD47 pKa = 4.04FFGTTYY53 pKa = 10.75SGLYY57 pKa = 9.7VNSNGNITFGAANTAYY73 pKa = 8.51QTSNLAGTSSRR84 pKa = 11.84MIAPFWGDD92 pKa = 3.18VNINSGGEE100 pKa = 4.09IYY102 pKa = 10.12WDD104 pKa = 3.44LDD106 pKa = 3.37AAAGTVTITWDD117 pKa = 3.22GVAPYY122 pKa = 9.69NGSGANSFQIVLTDD136 pKa = 3.2IGGGDD141 pKa = 3.52FTVEE145 pKa = 5.05FIYY148 pKa = 10.85EE149 pKa = 4.47DD150 pKa = 3.71IQWTTGYY157 pKa = 10.09SQVAQAGITDD167 pKa = 4.1GGANDD172 pKa = 5.44FILPGSGNATTMSNYY187 pKa = 6.77EE188 pKa = 4.13TYY190 pKa = 11.07DD191 pKa = 3.25FGTNDD196 pKa = 3.69PNGTTDD202 pKa = 4.04FFFVNGQPVFGDD214 pKa = 3.78GVVEE218 pKa = 4.43GTSGADD224 pKa = 3.46TIDD227 pKa = 2.98AAYY230 pKa = 10.83YY231 pKa = 10.48GDD233 pKa = 4.63PDD235 pKa = 4.74GDD237 pKa = 4.95LIDD240 pKa = 5.87DD241 pKa = 4.48GDD243 pKa = 4.18GTGPAGNEE251 pKa = 3.53DD252 pKa = 4.88LIYY255 pKa = 10.95GFDD258 pKa = 3.64GDD260 pKa = 5.03DD261 pKa = 4.22NIFAGDD267 pKa = 4.13GDD269 pKa = 3.92DD270 pKa = 4.2TIYY273 pKa = 11.3GGDD276 pKa = 3.48GTDD279 pKa = 3.77NIFGEE284 pKa = 4.96DD285 pKa = 4.02GDD287 pKa = 4.04DD288 pKa = 4.37TIYY291 pKa = 11.25GDD293 pKa = 3.81NGPTGVTEE301 pKa = 4.24DD302 pKa = 4.86LNWSAQGADD311 pKa = 3.51GASIAGGFTQTTGTMDD327 pKa = 3.39VNVSFSADD335 pKa = 2.89GNNAVFEE342 pKa = 4.41VATAEE347 pKa = 4.22TNYY350 pKa = 10.37VAGGEE355 pKa = 4.02PFAPNSSAYY364 pKa = 10.65VYY366 pKa = 11.2GNGDD370 pKa = 3.7GPTGTVTIDD379 pKa = 3.53FAAVPGSGMSDD390 pKa = 3.13EE391 pKa = 4.45VQNVAFRR398 pKa = 11.84LNDD401 pKa = 3.16IDD403 pKa = 5.12SYY405 pKa = 11.48AGNHH409 pKa = 6.18IDD411 pKa = 3.51VLTVNAYY418 pKa = 10.12DD419 pKa = 3.86EE420 pKa = 5.09NGDD423 pKa = 3.86PVAVTLTAAGNDD435 pKa = 3.65SVSGNTVTAGGTLDD449 pKa = 4.22NPQDD453 pKa = 3.66AQGSVLVEE461 pKa = 3.59IDD463 pKa = 4.38GPVAWFEE470 pKa = 4.31VIYY473 pKa = 11.1VNALTGTHH481 pKa = 7.55GINITDD487 pKa = 3.53VSFDD491 pKa = 3.96TIPASGAADD500 pKa = 3.79NIFGGLGDD508 pKa = 3.96DD509 pKa = 4.61TIHH512 pKa = 7.14GEE514 pKa = 4.51AGDD517 pKa = 3.82DD518 pKa = 4.39VIWGDD523 pKa = 3.91EE524 pKa = 4.26GSDD527 pKa = 3.51TLFGGAGDD535 pKa = 3.8DD536 pKa = 4.47TIWAGAGDD544 pKa = 4.67TVTGGAGDD552 pKa = 3.94DD553 pKa = 4.4MIWLDD558 pKa = 3.88AANALGGPGAVIDD571 pKa = 4.94IIGSEE576 pKa = 4.14DD577 pKa = 3.96GEE579 pKa = 4.74TGGDD583 pKa = 3.47TLNFAGLIDD592 pKa = 4.22WGTISYY598 pKa = 9.89TDD600 pKa = 4.03AEE602 pKa = 4.71SGSATLSDD610 pKa = 3.62GTTVTFSNIEE620 pKa = 3.74NLIICFTGGTAIDD633 pKa = 4.41TPSGPCPVEE642 pKa = 3.57HH643 pKa = 6.81LRR645 pKa = 11.84PGDD648 pKa = 3.61LVLTRR653 pKa = 11.84DD654 pKa = 4.33RR655 pKa = 11.84GPQPIRR661 pKa = 11.84WIGRR665 pKa = 11.84RR666 pKa = 11.84TVPGTGTFAPIRR678 pKa = 11.84FEE680 pKa = 4.44RR681 pKa = 11.84GAFGNVAPLMVSPQHH696 pKa = 6.25RR697 pKa = 11.84MVYY700 pKa = 9.77SGSRR704 pKa = 11.84ANLYY708 pKa = 10.41FDD710 pKa = 3.84TPEE713 pKa = 3.73VMVPAKK719 pKa = 10.5HH720 pKa = 6.64LVDD723 pKa = 3.36QRR725 pKa = 11.84LIRR728 pKa = 11.84QVEE731 pKa = 4.63VPTVTYY737 pKa = 8.48FHH739 pKa = 7.12IMLDD743 pKa = 3.2RR744 pKa = 11.84HH745 pKa = 5.22EE746 pKa = 4.46VLWGNGAPSEE756 pKa = 4.4SFHH759 pKa = 6.78PGALGLDD766 pKa = 3.48ALAAEE771 pKa = 4.58ARR773 pKa = 11.84EE774 pKa = 4.05EE775 pKa = 4.13LLTLFPDD782 pKa = 3.97LRR784 pKa = 11.84ADD786 pKa = 3.45PGVYY790 pKa = 10.14GKK792 pKa = 7.46TARR795 pKa = 11.84RR796 pKa = 11.84VLKK799 pKa = 10.5RR800 pKa = 11.84FEE802 pKa = 4.18ARR804 pKa = 11.84LLAAA808 pKa = 5.49
Molecular weight: 83.72 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A3VH24|A3VH24_9RHOB Uncharacterized protein OS=Maritimibacter alkaliphilus HTCC2654 OX=314271 GN=RB2654_14875 PE=4 SV=1
MM1 pKa = 7.4RR2 pKa = 11.84RR3 pKa = 11.84GRR5 pKa = 11.84IGSTGSAIFRR15 pKa = 11.84RR16 pKa = 11.84PGSTATIRR24 pKa = 11.84VAGSMRR30 pKa = 11.84KK31 pKa = 9.34PGG33 pKa = 3.3
MM1 pKa = 7.4RR2 pKa = 11.84RR3 pKa = 11.84GRR5 pKa = 11.84IGSTGSAIFRR15 pKa = 11.84RR16 pKa = 11.84PGSTATIRR24 pKa = 11.84VAGSMRR30 pKa = 11.84KK31 pKa = 9.34PGG33 pKa = 3.3
Molecular weight: 3.49 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1363242 |
15 |
7434 |
290.5 |
31.54 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.122 ± 0.052 | 0.797 ± 0.012 |
6.541 ± 0.047 | 6.1 ± 0.037 |
3.794 ± 0.023 | 8.895 ± 0.041 |
2.035 ± 0.021 | 5.128 ± 0.028 |
3.186 ± 0.034 | 9.53 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.835 ± 0.021 | 2.574 ± 0.023 |
5.091 ± 0.031 | 2.86 ± 0.02 |
6.711 ± 0.049 | 4.935 ± 0.031 |
5.723 ± 0.039 | 7.523 ± 0.03 |
1.388 ± 0.017 | 2.233 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
