
Pseudocercospora fijiensis (strain CIRAD86) (Black leaf streak disease fungus) (Mycosphaerella fijiensis)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; dothideomyceta; Dothideomycetes; Dothideomycetidae; Mycosphaerellales; Mycosphaerellaceae; Pseudocercospora; Pseudocercospora fijiensis
Average proteome isoelectric point is 6.79
Get precalculated fractions of proteins
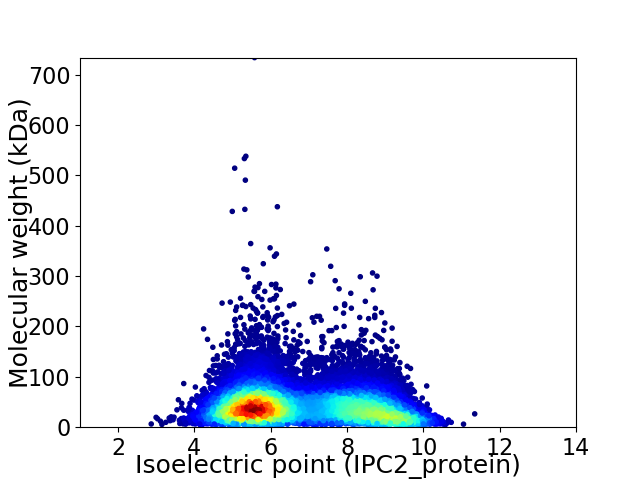
Virtual 2D-PAGE plot for 13062 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|M3B4A1|M3B4A1_PSEFD PPM-type phosphatase domain-containing protein OS=Pseudocercospora fijiensis (strain CIRAD86) OX=383855 GN=MYCFIDRAFT_133445 PE=3 SV=1
NN1 pKa = 7.92DD2 pKa = 4.59CGDD5 pKa = 3.51SSFVKK10 pKa = 9.31VTMSNRR16 pKa = 11.84PLVADD21 pKa = 4.38CQQLVANLAGNQEE34 pKa = 3.97WRR36 pKa = 11.84VTTSGVQVAVYY47 pKa = 7.25QTCAFNAVSFGGDD60 pKa = 3.66SIIGNADD67 pKa = 3.63VIDD70 pKa = 5.67LINDD74 pKa = 4.27SISKK78 pKa = 10.73FEE80 pKa = 4.69DD81 pKa = 2.85NGYY84 pKa = 10.76VGAQGG89 pKa = 3.31
NN1 pKa = 7.92DD2 pKa = 4.59CGDD5 pKa = 3.51SSFVKK10 pKa = 9.31VTMSNRR16 pKa = 11.84PLVADD21 pKa = 4.38CQQLVANLAGNQEE34 pKa = 3.97WRR36 pKa = 11.84VTTSGVQVAVYY47 pKa = 7.25QTCAFNAVSFGGDD60 pKa = 3.66SIIGNADD67 pKa = 3.63VIDD70 pKa = 5.67LINDD74 pKa = 4.27SISKK78 pKa = 10.73FEE80 pKa = 4.69DD81 pKa = 2.85NGYY84 pKa = 10.76VGAQGG89 pKa = 3.31
Molecular weight: 9.38 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|M3AH58|M3AH58_PSEFD Uncharacterized protein OS=Pseudocercospora fijiensis (strain CIRAD86) OX=383855 GN=MYCFIDRAFT_214540 PE=4 SV=1
MM1 pKa = 7.44VLLLLLLIQRR11 pKa = 11.84LLQRR15 pKa = 11.84LLQRR19 pKa = 11.84LLQLQIPVVQVLVVLVLLLLLLIQRR44 pKa = 11.84LLQLQILVVQALVVQVLLLLLLIQRR69 pKa = 11.84LLQLQILVAQALMAQALVVQALVVQALVVQVLVVQALVVQALVVQALVVQALVVQVLVVQALVVQALVVRR139 pKa = 11.84APVVRR144 pKa = 11.84APVVRR149 pKa = 11.84APVVQALVVQALVVQALVVQALVVQALVVQALVVQAPVVRR189 pKa = 11.84ALVVQALVVQALQIPKK205 pKa = 9.68HH206 pKa = 5.51LHH208 pKa = 4.78QQSQLPNRR216 pKa = 11.84KK217 pKa = 8.7IPKK220 pKa = 8.6HH221 pKa = 4.39LHH223 pKa = 5.5RR224 pKa = 11.84LRR226 pKa = 11.84RR227 pKa = 11.84WAPPLMIMLRR237 pKa = 11.84FTAA240 pKa = 4.71
MM1 pKa = 7.44VLLLLLLIQRR11 pKa = 11.84LLQRR15 pKa = 11.84LLQRR19 pKa = 11.84LLQLQIPVVQVLVVLVLLLLLLIQRR44 pKa = 11.84LLQLQILVVQALVVQVLLLLLLIQRR69 pKa = 11.84LLQLQILVAQALMAQALVVQALVVQALVVQVLVVQALVVQALVVQALVVQALVVQVLVVQALVVQALVVRR139 pKa = 11.84APVVRR144 pKa = 11.84APVVRR149 pKa = 11.84APVVQALVVQALVVQALVVQALVVQALVVQALVVQAPVVRR189 pKa = 11.84ALVVQALVVQALQIPKK205 pKa = 9.68HH206 pKa = 5.51LHH208 pKa = 4.78QQSQLPNRR216 pKa = 11.84KK217 pKa = 8.7IPKK220 pKa = 8.6HH221 pKa = 4.39LHH223 pKa = 5.5RR224 pKa = 11.84LRR226 pKa = 11.84RR227 pKa = 11.84WAPPLMIMLRR237 pKa = 11.84FTAA240 pKa = 4.71
Molecular weight: 26.45 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
5586011 |
49 |
6685 |
427.7 |
47.46 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.921 ± 0.017 | 1.436 ± 0.01 |
5.559 ± 0.016 | 6.106 ± 0.021 |
3.665 ± 0.014 | 6.722 ± 0.02 |
2.63 ± 0.011 | 4.83 ± 0.014 |
4.959 ± 0.021 | 8.839 ± 0.024 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.236 ± 0.008 | 3.618 ± 0.013 |
5.835 ± 0.02 | 4.17 ± 0.015 |
6.401 ± 0.022 | 8.04 ± 0.023 |
5.89 ± 0.017 | 5.877 ± 0.015 |
1.523 ± 0.008 | 2.744 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
