
Human papillomavirus 158
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Gammapapillomavirus; Gammapapillomavirus 12
Average proteome isoelectric point is 6.08
Get precalculated fractions of proteins
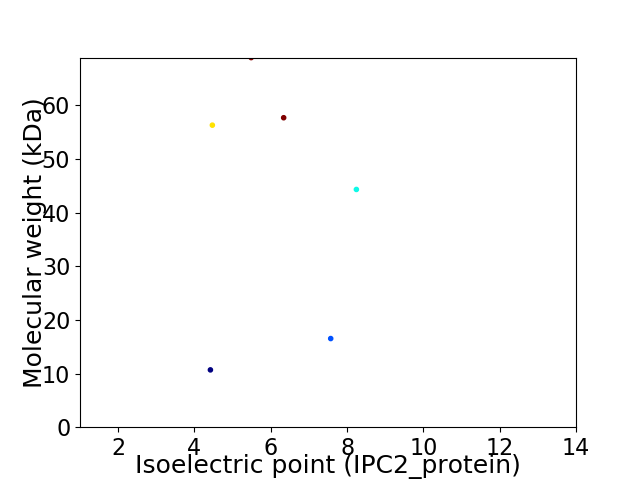
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A166FKE6|A0A166FKE6_9PAPI Replication protein E1 OS=Human papillomavirus 158 OX=2259343 GN=E1 PE=3 SV=1
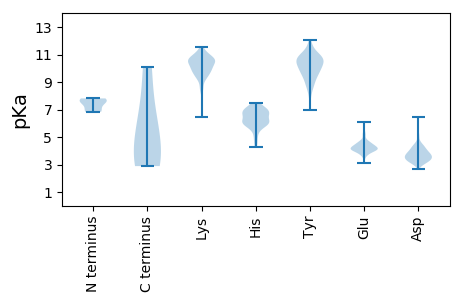
MM1 pKa = 7.77RR2 pKa = 11.84GAEE5 pKa = 3.94PTIADD10 pKa = 4.24IEE12 pKa = 4.52LDD14 pKa = 3.97LNDD17 pKa = 5.18LVIPSNLLSEE27 pKa = 5.25EE28 pKa = 4.11VLQDD32 pKa = 3.53EE33 pKa = 4.5NSEE36 pKa = 4.29EE37 pKa = 4.15EE38 pKa = 4.32LVPYY42 pKa = 10.29HH43 pKa = 7.45IDD45 pKa = 3.45TCCVQCQTGIRR56 pKa = 11.84LTIYY60 pKa = 10.21AVEE63 pKa = 4.04FALRR67 pKa = 11.84LLEE70 pKa = 5.15SLLLDD75 pKa = 3.95EE76 pKa = 6.08KK77 pKa = 11.4LLLCCPGCARR87 pKa = 11.84QSRR90 pKa = 11.84RR91 pKa = 11.84NGRR94 pKa = 11.84PP95 pKa = 2.88
MM1 pKa = 7.77RR2 pKa = 11.84GAEE5 pKa = 3.94PTIADD10 pKa = 4.24IEE12 pKa = 4.52LDD14 pKa = 3.97LNDD17 pKa = 5.18LVIPSNLLSEE27 pKa = 5.25EE28 pKa = 4.11VLQDD32 pKa = 3.53EE33 pKa = 4.5NSEE36 pKa = 4.29EE37 pKa = 4.15EE38 pKa = 4.32LVPYY42 pKa = 10.29HH43 pKa = 7.45IDD45 pKa = 3.45TCCVQCQTGIRR56 pKa = 11.84LTIYY60 pKa = 10.21AVEE63 pKa = 4.04FALRR67 pKa = 11.84LLEE70 pKa = 5.15SLLLDD75 pKa = 3.95EE76 pKa = 6.08KK77 pKa = 11.4LLLCCPGCARR87 pKa = 11.84QSRR90 pKa = 11.84RR91 pKa = 11.84NGRR94 pKa = 11.84PP95 pKa = 2.88
Molecular weight: 10.69 kDa
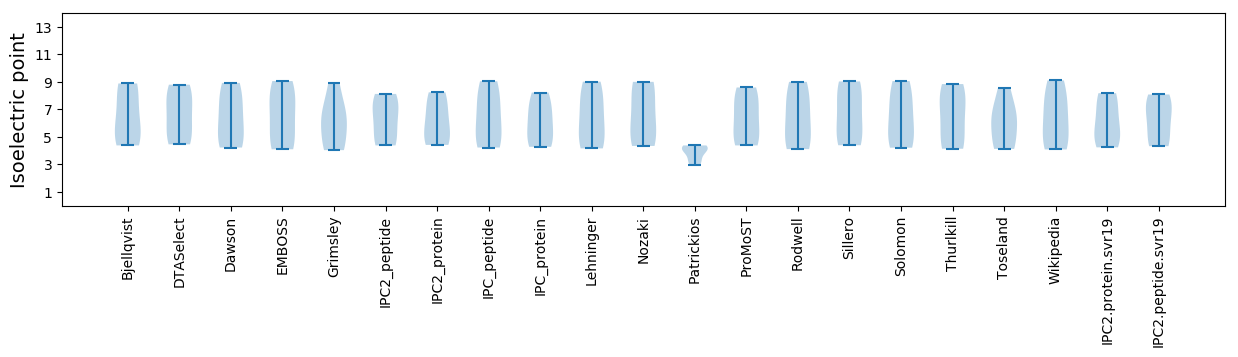
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A166FKG3|A0A166FKG3_9PAPI Minor capsid protein L2 OS=Human papillomavirus 158 OX=2259343 GN=L2 PE=3 SV=1
MM1 pKa = 6.9EE2 pKa = 4.33TLAEE6 pKa = 4.24RR7 pKa = 11.84FDD9 pKa = 3.78VLQEE13 pKa = 4.36EE14 pKa = 4.49ILKK17 pKa = 10.38HH18 pKa = 5.92IEE20 pKa = 4.09SGDD23 pKa = 3.48TSLQSHH29 pKa = 6.44IDD31 pKa = 3.08YY32 pKa = 9.98WEE34 pKa = 3.86NVRR37 pKa = 11.84KK38 pKa = 9.83EE39 pKa = 4.05NAIMYY44 pKa = 6.94YY45 pKa = 10.38ARR47 pKa = 11.84KK48 pKa = 9.5QGLSTLSLQPLPQLAVSEE66 pKa = 4.62YY67 pKa = 9.83NAKK70 pKa = 9.9QAIQISLTLQSLLKK84 pKa = 10.86SPFGNEE90 pKa = 3.1AWTLPEE96 pKa = 4.19VSAEE100 pKa = 4.51LINTPPQNVLKK111 pKa = 10.96KK112 pKa = 10.14NGYY115 pKa = 9.25DD116 pKa = 3.48VEE118 pKa = 3.93IWYY121 pKa = 10.0DD122 pKa = 3.26NDD124 pKa = 3.11RR125 pKa = 11.84RR126 pKa = 11.84NSMVYY131 pKa = 9.52TNWNSLYY138 pKa = 10.56YY139 pKa = 9.7QDD141 pKa = 6.16HH142 pKa = 6.02EE143 pKa = 4.77EE144 pKa = 4.08KK145 pKa = 8.47WHH147 pKa = 6.21KK148 pKa = 9.73VTGQVDD154 pKa = 3.54YY155 pKa = 10.86DD156 pKa = 3.47GCYY159 pKa = 8.83FTDD162 pKa = 3.46HH163 pKa = 6.27TGEE166 pKa = 3.68RR167 pKa = 11.84AYY169 pKa = 10.75FILFITDD176 pKa = 3.52ASRR179 pKa = 11.84YY180 pKa = 8.86SEE182 pKa = 4.04SGTWTVHH189 pKa = 5.43YY190 pKa = 8.69KK191 pKa = 9.55QQVISSSVVSSSNYY205 pKa = 9.56SAYY208 pKa = 9.98EE209 pKa = 4.03DD210 pKa = 4.55PLPGPSSNTEE220 pKa = 4.08TPSKK224 pKa = 9.77NHH226 pKa = 5.33LRR228 pKa = 11.84RR229 pKa = 11.84RR230 pKa = 11.84PEE232 pKa = 3.82EE233 pKa = 4.01TNLPSSTTPSTSPRR247 pKa = 11.84SLRR250 pKa = 11.84VRR252 pKa = 11.84RR253 pKa = 11.84GRR255 pKa = 11.84RR256 pKa = 11.84EE257 pKa = 3.66QGEE260 pKa = 4.3SGTRR264 pKa = 11.84GEE266 pKa = 4.48TPAKK270 pKa = 9.52RR271 pKa = 11.84RR272 pKa = 11.84RR273 pKa = 11.84GGGSGSAPTPSEE285 pKa = 3.65VGSRR289 pKa = 11.84HH290 pKa = 4.97RR291 pKa = 11.84QVEE294 pKa = 4.27RR295 pKa = 11.84KK296 pKa = 8.97GLSRR300 pKa = 11.84LGVLQEE306 pKa = 4.06EE307 pKa = 4.7ARR309 pKa = 11.84DD310 pKa = 3.63PPIIVVQGCANGLKK324 pKa = 9.96CWRR327 pKa = 11.84NRR329 pKa = 11.84KK330 pKa = 8.36SNKK333 pKa = 9.03HH334 pKa = 5.78AGSFLAMSTVWNWVGDD350 pKa = 3.6TSANHH355 pKa = 5.83SRR357 pKa = 11.84LIIAFANTAQRR368 pKa = 11.84EE369 pKa = 4.49HH370 pKa = 6.15FVKK373 pKa = 10.59NHH375 pKa = 6.81LFPKK379 pKa = 9.84HH380 pKa = 4.31ATYY383 pKa = 10.9YY384 pKa = 9.37YY385 pKa = 10.91GSLGGLL391 pKa = 3.54
MM1 pKa = 6.9EE2 pKa = 4.33TLAEE6 pKa = 4.24RR7 pKa = 11.84FDD9 pKa = 3.78VLQEE13 pKa = 4.36EE14 pKa = 4.49ILKK17 pKa = 10.38HH18 pKa = 5.92IEE20 pKa = 4.09SGDD23 pKa = 3.48TSLQSHH29 pKa = 6.44IDD31 pKa = 3.08YY32 pKa = 9.98WEE34 pKa = 3.86NVRR37 pKa = 11.84KK38 pKa = 9.83EE39 pKa = 4.05NAIMYY44 pKa = 6.94YY45 pKa = 10.38ARR47 pKa = 11.84KK48 pKa = 9.5QGLSTLSLQPLPQLAVSEE66 pKa = 4.62YY67 pKa = 9.83NAKK70 pKa = 9.9QAIQISLTLQSLLKK84 pKa = 10.86SPFGNEE90 pKa = 3.1AWTLPEE96 pKa = 4.19VSAEE100 pKa = 4.51LINTPPQNVLKK111 pKa = 10.96KK112 pKa = 10.14NGYY115 pKa = 9.25DD116 pKa = 3.48VEE118 pKa = 3.93IWYY121 pKa = 10.0DD122 pKa = 3.26NDD124 pKa = 3.11RR125 pKa = 11.84RR126 pKa = 11.84NSMVYY131 pKa = 9.52TNWNSLYY138 pKa = 10.56YY139 pKa = 9.7QDD141 pKa = 6.16HH142 pKa = 6.02EE143 pKa = 4.77EE144 pKa = 4.08KK145 pKa = 8.47WHH147 pKa = 6.21KK148 pKa = 9.73VTGQVDD154 pKa = 3.54YY155 pKa = 10.86DD156 pKa = 3.47GCYY159 pKa = 8.83FTDD162 pKa = 3.46HH163 pKa = 6.27TGEE166 pKa = 3.68RR167 pKa = 11.84AYY169 pKa = 10.75FILFITDD176 pKa = 3.52ASRR179 pKa = 11.84YY180 pKa = 8.86SEE182 pKa = 4.04SGTWTVHH189 pKa = 5.43YY190 pKa = 8.69KK191 pKa = 9.55QQVISSSVVSSSNYY205 pKa = 9.56SAYY208 pKa = 9.98EE209 pKa = 4.03DD210 pKa = 4.55PLPGPSSNTEE220 pKa = 4.08TPSKK224 pKa = 9.77NHH226 pKa = 5.33LRR228 pKa = 11.84RR229 pKa = 11.84RR230 pKa = 11.84PEE232 pKa = 3.82EE233 pKa = 4.01TNLPSSTTPSTSPRR247 pKa = 11.84SLRR250 pKa = 11.84VRR252 pKa = 11.84RR253 pKa = 11.84GRR255 pKa = 11.84RR256 pKa = 11.84EE257 pKa = 3.66QGEE260 pKa = 4.3SGTRR264 pKa = 11.84GEE266 pKa = 4.48TPAKK270 pKa = 9.52RR271 pKa = 11.84RR272 pKa = 11.84RR273 pKa = 11.84GGGSGSAPTPSEE285 pKa = 3.65VGSRR289 pKa = 11.84HH290 pKa = 4.97RR291 pKa = 11.84QVEE294 pKa = 4.27RR295 pKa = 11.84KK296 pKa = 8.97GLSRR300 pKa = 11.84LGVLQEE306 pKa = 4.06EE307 pKa = 4.7ARR309 pKa = 11.84DD310 pKa = 3.63PPIIVVQGCANGLKK324 pKa = 9.96CWRR327 pKa = 11.84NRR329 pKa = 11.84KK330 pKa = 8.36SNKK333 pKa = 9.03HH334 pKa = 5.78AGSFLAMSTVWNWVGDD350 pKa = 3.6TSANHH355 pKa = 5.83SRR357 pKa = 11.84LIIAFANTAQRR368 pKa = 11.84EE369 pKa = 4.49HH370 pKa = 6.15FVKK373 pKa = 10.59NHH375 pKa = 6.81LFPKK379 pKa = 9.84HH380 pKa = 4.31ATYY383 pKa = 10.9YY384 pKa = 9.37YY385 pKa = 10.91GSLGGLL391 pKa = 3.54
Molecular weight: 44.33 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2258 |
95 |
600 |
376.3 |
42.42 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.624 ± 0.314 | 2.17 ± 0.897 |
6.466 ± 0.799 | 5.979 ± 0.607 |
4.739 ± 0.636 | 5.846 ± 0.78 |
1.993 ± 0.282 | 5.093 ± 0.809 |
5.093 ± 0.918 | 9.123 ± 0.733 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.417 ± 0.192 | 5.049 ± 0.514 |
5.624 ± 0.903 | 4.562 ± 0.356 |
5.713 ± 0.654 | 7.795 ± 0.641 |
6.466 ± 0.605 | 6.599 ± 0.458 |
1.329 ± 0.345 | 3.322 ± 0.4 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
