
Vannielia litorea
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Vannielia
Average proteome isoelectric point is 6.19
Get precalculated fractions of proteins
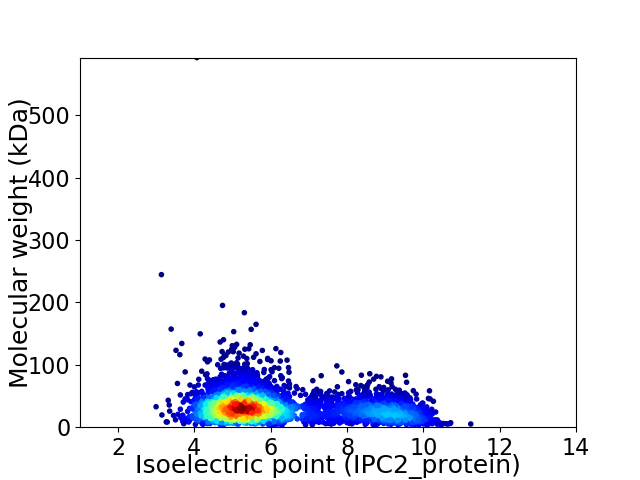
Virtual 2D-PAGE plot for 4018 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1N6E734|A0A1N6E734_9RHOB Uncharacterized protein OS=Vannielia litorea OX=1217970 GN=SAMN05444002_0409 PE=4 SV=1
MM1 pKa = 7.4LLDD4 pKa = 4.08EE5 pKa = 4.68KK6 pKa = 11.35SEE8 pKa = 4.33IIAHH12 pKa = 6.62FIGLFQLGSEE22 pKa = 4.47AARR25 pKa = 11.84MRR27 pKa = 11.84EE28 pKa = 3.92EE29 pKa = 4.03FEE31 pKa = 4.07EE32 pKa = 4.5FQALKK37 pKa = 10.79NADD40 pKa = 3.84PEE42 pKa = 4.55FGSLLNITINLFSDD56 pKa = 4.05YY57 pKa = 10.57EE58 pKa = 4.24LGAFTPDD65 pKa = 2.83IQGPLYY71 pKa = 10.52FPVLPGVPGLPYY83 pKa = 10.77LPFAGPAAEE92 pKa = 4.57HH93 pKa = 6.67LWPVLPGEE101 pKa = 4.08LAEE104 pKa = 5.42LPIYY108 pKa = 9.55WSPPGYY114 pKa = 10.75LLAKK118 pKa = 9.81LQFLIPPPSSVATITVQYY136 pKa = 11.03NFLRR140 pKa = 11.84DD141 pKa = 3.24YY142 pKa = 11.03DD143 pKa = 4.07YY144 pKa = 11.82ASTSDD149 pKa = 3.64AEE151 pKa = 4.3MSFVDD156 pKa = 4.25PVAFEE161 pKa = 4.6VPLSSLQATATALQPFSAPVMPEE184 pKa = 3.57DD185 pKa = 3.48EE186 pKa = 4.22AAIAEE191 pKa = 4.42SGRR194 pKa = 11.84EE195 pKa = 3.72VSEE198 pKa = 5.1DD199 pKa = 2.8IRR201 pKa = 11.84ALEE204 pKa = 4.23SEE206 pKa = 4.65GGPGAADD213 pKa = 3.45GAVTFVAFGEE223 pKa = 4.4ATATPTLNGLAVEE236 pKa = 4.58EE237 pKa = 4.28MPKK240 pKa = 10.69LSDD243 pKa = 3.62YY244 pKa = 11.33SDD246 pKa = 3.43TFAEE250 pKa = 4.5PEE252 pKa = 4.13PEE254 pKa = 4.11PEE256 pKa = 4.15EE257 pKa = 4.13EE258 pKa = 4.33VPEE261 pKa = 5.12SPFEE265 pKa = 4.49DD266 pKa = 3.49NRR268 pKa = 11.84DD269 pKa = 3.44EE270 pKa = 5.42AEE272 pKa = 4.24EE273 pKa = 4.51DD274 pKa = 3.47DD275 pKa = 4.56HH276 pKa = 9.11PEE278 pKa = 4.22GQHH281 pKa = 4.08VTMGEE286 pKa = 3.85NLLINQANVVMNWLDD301 pKa = 3.41APVFVVMGDD310 pKa = 3.5VVVANAISQINVWNDD325 pKa = 2.85FDD327 pKa = 4.32KK328 pKa = 11.53VSGVSSLTEE337 pKa = 4.04AGSTRR342 pKa = 11.84SVNASSIEE350 pKa = 4.1HH351 pKa = 5.6VANPVSVGEE360 pKa = 4.13GAYY363 pKa = 10.49GPGQAIVTRR372 pKa = 11.84IEE374 pKa = 4.19GNVINYY380 pKa = 7.8NHH382 pKa = 6.47IQQFNFAAEE391 pKa = 4.19GDD393 pKa = 4.18VLSLLFNASEE403 pKa = 4.42TFIQTGGNTLFNIADD418 pKa = 3.52ILEE421 pKa = 4.51LGFGYY426 pKa = 10.67DD427 pKa = 4.47LIVIGGNMIDD437 pKa = 3.74VTLLTQMNVLLDD449 pKa = 3.96ADD451 pKa = 3.74SAEE454 pKa = 4.19IEE456 pKa = 4.48GEE458 pKa = 4.02FGGTMDD464 pKa = 3.85TSGNLLLNYY473 pKa = 10.11AAITEE478 pKa = 4.37VGVDD482 pKa = 3.23TTVAITEE489 pKa = 4.61AYY491 pKa = 9.97SKK493 pKa = 10.77AANKK497 pKa = 9.73VIEE500 pKa = 4.56GGEE503 pKa = 3.97PGGDD507 pKa = 2.93ILGDD511 pKa = 3.51PAYY514 pKa = 10.7LGTPVLRR521 pKa = 11.84VLYY524 pKa = 10.43IEE526 pKa = 5.33GDD528 pKa = 3.81YY529 pKa = 11.35LDD531 pKa = 3.73VQVIEE536 pKa = 4.19QVNVLGDD543 pKa = 3.53VDD545 pKa = 4.27QVALAAEE552 pKa = 4.37TLLSAEE558 pKa = 4.35GADD561 pKa = 3.52VTVLTGGNEE570 pKa = 4.22LVNIASVTDD579 pKa = 3.54HH580 pKa = 6.69GVDD583 pKa = 3.32STVYY587 pKa = 9.4TGGEE591 pKa = 4.13AYY593 pKa = 10.45SDD595 pKa = 3.34AMLYY599 pKa = 9.39QAEE602 pKa = 5.09FISNDD607 pKa = 3.47DD608 pKa = 3.75PFALASSDD616 pKa = 3.57NLASEE621 pKa = 4.74AVLFLADD628 pKa = 5.55DD629 pKa = 4.12MLSDD633 pKa = 3.74TSDD636 pKa = 3.5GPGAGLGQLVPEE648 pKa = 4.52EE649 pKa = 4.46APVDD653 pKa = 3.87VMQTLVAA660 pKa = 4.55
MM1 pKa = 7.4LLDD4 pKa = 4.08EE5 pKa = 4.68KK6 pKa = 11.35SEE8 pKa = 4.33IIAHH12 pKa = 6.62FIGLFQLGSEE22 pKa = 4.47AARR25 pKa = 11.84MRR27 pKa = 11.84EE28 pKa = 3.92EE29 pKa = 4.03FEE31 pKa = 4.07EE32 pKa = 4.5FQALKK37 pKa = 10.79NADD40 pKa = 3.84PEE42 pKa = 4.55FGSLLNITINLFSDD56 pKa = 4.05YY57 pKa = 10.57EE58 pKa = 4.24LGAFTPDD65 pKa = 2.83IQGPLYY71 pKa = 10.52FPVLPGVPGLPYY83 pKa = 10.77LPFAGPAAEE92 pKa = 4.57HH93 pKa = 6.67LWPVLPGEE101 pKa = 4.08LAEE104 pKa = 5.42LPIYY108 pKa = 9.55WSPPGYY114 pKa = 10.75LLAKK118 pKa = 9.81LQFLIPPPSSVATITVQYY136 pKa = 11.03NFLRR140 pKa = 11.84DD141 pKa = 3.24YY142 pKa = 11.03DD143 pKa = 4.07YY144 pKa = 11.82ASTSDD149 pKa = 3.64AEE151 pKa = 4.3MSFVDD156 pKa = 4.25PVAFEE161 pKa = 4.6VPLSSLQATATALQPFSAPVMPEE184 pKa = 3.57DD185 pKa = 3.48EE186 pKa = 4.22AAIAEE191 pKa = 4.42SGRR194 pKa = 11.84EE195 pKa = 3.72VSEE198 pKa = 5.1DD199 pKa = 2.8IRR201 pKa = 11.84ALEE204 pKa = 4.23SEE206 pKa = 4.65GGPGAADD213 pKa = 3.45GAVTFVAFGEE223 pKa = 4.4ATATPTLNGLAVEE236 pKa = 4.58EE237 pKa = 4.28MPKK240 pKa = 10.69LSDD243 pKa = 3.62YY244 pKa = 11.33SDD246 pKa = 3.43TFAEE250 pKa = 4.5PEE252 pKa = 4.13PEE254 pKa = 4.11PEE256 pKa = 4.15EE257 pKa = 4.13EE258 pKa = 4.33VPEE261 pKa = 5.12SPFEE265 pKa = 4.49DD266 pKa = 3.49NRR268 pKa = 11.84DD269 pKa = 3.44EE270 pKa = 5.42AEE272 pKa = 4.24EE273 pKa = 4.51DD274 pKa = 3.47DD275 pKa = 4.56HH276 pKa = 9.11PEE278 pKa = 4.22GQHH281 pKa = 4.08VTMGEE286 pKa = 3.85NLLINQANVVMNWLDD301 pKa = 3.41APVFVVMGDD310 pKa = 3.5VVVANAISQINVWNDD325 pKa = 2.85FDD327 pKa = 4.32KK328 pKa = 11.53VSGVSSLTEE337 pKa = 4.04AGSTRR342 pKa = 11.84SVNASSIEE350 pKa = 4.1HH351 pKa = 5.6VANPVSVGEE360 pKa = 4.13GAYY363 pKa = 10.49GPGQAIVTRR372 pKa = 11.84IEE374 pKa = 4.19GNVINYY380 pKa = 7.8NHH382 pKa = 6.47IQQFNFAAEE391 pKa = 4.19GDD393 pKa = 4.18VLSLLFNASEE403 pKa = 4.42TFIQTGGNTLFNIADD418 pKa = 3.52ILEE421 pKa = 4.51LGFGYY426 pKa = 10.67DD427 pKa = 4.47LIVIGGNMIDD437 pKa = 3.74VTLLTQMNVLLDD449 pKa = 3.96ADD451 pKa = 3.74SAEE454 pKa = 4.19IEE456 pKa = 4.48GEE458 pKa = 4.02FGGTMDD464 pKa = 3.85TSGNLLLNYY473 pKa = 10.11AAITEE478 pKa = 4.37VGVDD482 pKa = 3.23TTVAITEE489 pKa = 4.61AYY491 pKa = 9.97SKK493 pKa = 10.77AANKK497 pKa = 9.73VIEE500 pKa = 4.56GGEE503 pKa = 3.97PGGDD507 pKa = 2.93ILGDD511 pKa = 3.51PAYY514 pKa = 10.7LGTPVLRR521 pKa = 11.84VLYY524 pKa = 10.43IEE526 pKa = 5.33GDD528 pKa = 3.81YY529 pKa = 11.35LDD531 pKa = 3.73VQVIEE536 pKa = 4.19QVNVLGDD543 pKa = 3.53VDD545 pKa = 4.27QVALAAEE552 pKa = 4.37TLLSAEE558 pKa = 4.35GADD561 pKa = 3.52VTVLTGGNEE570 pKa = 4.22LVNIASVTDD579 pKa = 3.54HH580 pKa = 6.69GVDD583 pKa = 3.32STVYY587 pKa = 9.4TGGEE591 pKa = 4.13AYY593 pKa = 10.45SDD595 pKa = 3.34AMLYY599 pKa = 9.39QAEE602 pKa = 5.09FISNDD607 pKa = 3.47DD608 pKa = 3.75PFALASSDD616 pKa = 3.57NLASEE621 pKa = 4.74AVLFLADD628 pKa = 5.55DD629 pKa = 4.12MLSDD633 pKa = 3.74TSDD636 pKa = 3.5GPGAGLGQLVPEE648 pKa = 4.52EE649 pKa = 4.46APVDD653 pKa = 3.87VMQTLVAA660 pKa = 4.55
Molecular weight: 70.11 kDa
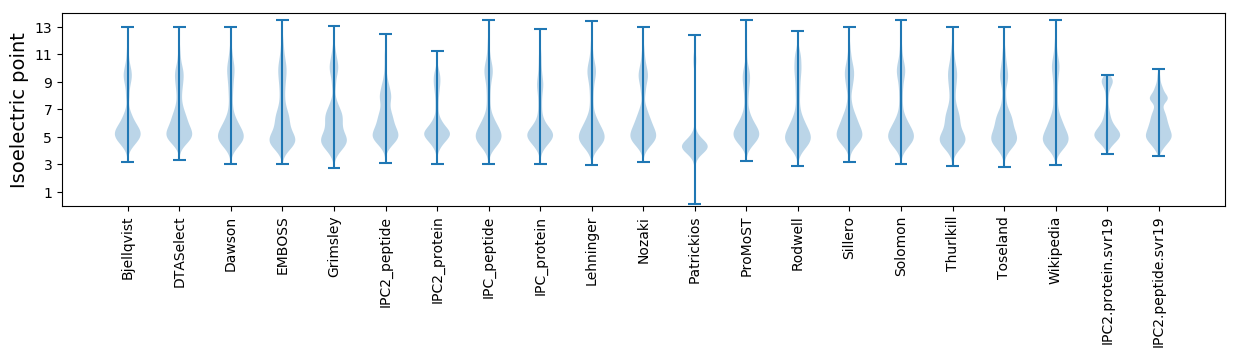
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1N6ICQ0|A0A1N6ICQ0_9RHOB Putative membrane protein insertion efficiency factor OS=Vannielia litorea OX=1217970 GN=SAMN05444002_3711 PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
Molecular weight: 5.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1242426 |
29 |
5927 |
309.2 |
33.36 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.271 ± 0.064 | 0.895 ± 0.014 |
5.439 ± 0.035 | 6.466 ± 0.04 |
3.642 ± 0.029 | 9.158 ± 0.043 |
2.003 ± 0.02 | 4.715 ± 0.032 |
2.936 ± 0.03 | 10.192 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.709 ± 0.021 | 2.355 ± 0.03 |
5.347 ± 0.039 | 2.89 ± 0.019 |
6.821 ± 0.044 | 4.884 ± 0.032 |
5.368 ± 0.044 | 7.329 ± 0.034 |
1.452 ± 0.018 | 2.131 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
