
Pacific flying fox faeces associated circular DNA virus-14
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 7.89
Get precalculated fractions of proteins
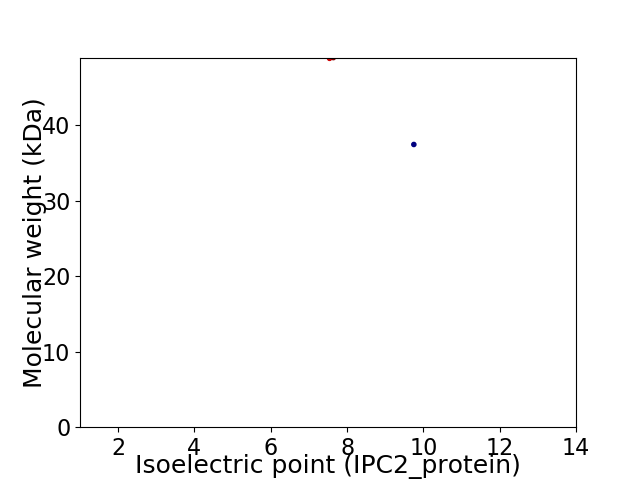
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A140CTW2|A0A140CTW2_9VIRU Replication-associated protein OS=Pacific flying fox faeces associated circular DNA virus-14 OX=1796009 PE=3 SV=1
MM1 pKa = 7.49NSVCTLSHH9 pKa = 7.12RR10 pKa = 11.84STSLLSATTTVTDD23 pKa = 3.38PHH25 pKa = 5.76TTTTEE30 pKa = 3.8TTRR33 pKa = 11.84PDD35 pKa = 2.99IVIRR39 pKa = 11.84PDD41 pKa = 3.38TLISANRR48 pKa = 11.84NQDD51 pKa = 3.48DD52 pKa = 3.98EE53 pKa = 4.87TGLEE57 pKa = 4.63PIPDD61 pKa = 3.28KK62 pKa = 11.2NFRR65 pKa = 11.84LNCKK69 pKa = 9.83AVFLTYY75 pKa = 8.8PQCALEE81 pKa = 4.47PEE83 pKa = 4.85HH84 pKa = 7.2VLQTILACNKK94 pKa = 8.17TKK96 pKa = 10.21HH97 pKa = 6.28CSVIVAQEE105 pKa = 3.7HH106 pKa = 6.38HH107 pKa = 6.91KK108 pKa = 11.07DD109 pKa = 3.8GNKK112 pKa = 9.62HH113 pKa = 4.28LHH115 pKa = 6.27VYY117 pKa = 10.92LEE119 pKa = 4.23FPKK122 pKa = 10.8KK123 pKa = 9.92IDD125 pKa = 3.17IKK127 pKa = 7.71TARR130 pKa = 11.84YY131 pKa = 8.76FNYY134 pKa = 8.73ITGKK138 pKa = 8.93QGNYY142 pKa = 7.83QKK144 pKa = 11.1VKK146 pKa = 10.6VRR148 pKa = 11.84GACIKK153 pKa = 9.32YY154 pKa = 8.2VCKK157 pKa = 10.09EE158 pKa = 3.72NRR160 pKa = 11.84YY161 pKa = 8.82VAHH164 pKa = 6.58QIDD167 pKa = 4.6VPTIIQNYY175 pKa = 7.86EE176 pKa = 3.58KK177 pKa = 10.46QQEE180 pKa = 4.39KK181 pKa = 10.49KK182 pKa = 10.42KK183 pKa = 10.87SRR185 pKa = 11.84NQAKK189 pKa = 9.95NKK191 pKa = 9.68RR192 pKa = 11.84VYY194 pKa = 10.74EE195 pKa = 4.21LIKK198 pKa = 10.39EE199 pKa = 4.29GKK201 pKa = 9.85KK202 pKa = 10.23YY203 pKa = 10.91EE204 pKa = 4.25EE205 pKa = 4.89LLLDD209 pKa = 4.23CTTGPYY215 pKa = 10.3CLLHH219 pKa = 6.12GNKK222 pKa = 8.76IKK224 pKa = 11.18NMITDD229 pKa = 4.16FEE231 pKa = 4.69EE232 pKa = 4.21IAMKK236 pKa = 9.79KK237 pKa = 8.6QRR239 pKa = 11.84IADD242 pKa = 4.11RR243 pKa = 11.84PAYY246 pKa = 10.12LHH248 pKa = 6.96LILPGKK254 pKa = 10.32DD255 pKa = 2.8IDD257 pKa = 4.75LLAKK261 pKa = 10.38KK262 pKa = 9.59PFKK265 pKa = 10.91SPQFWIFGPPNVGKK279 pKa = 8.97TSMLQSLFDD288 pKa = 4.12AGLKK292 pKa = 10.45GFEE295 pKa = 4.06IPINNDD301 pKa = 2.74FARR304 pKa = 11.84WDD306 pKa = 3.63DD307 pKa = 3.66EE308 pKa = 4.92LFDD311 pKa = 3.92FAYY314 pKa = 9.71IDD316 pKa = 3.69EE317 pKa = 5.06FKK319 pKa = 11.18GQLTIQFLNSFLQGSRR335 pKa = 11.84MALPGKK341 pKa = 9.61YY342 pKa = 10.31VIGGKK347 pKa = 9.3IKK349 pKa = 10.74NKK351 pKa = 10.21NIPTFILSNYY361 pKa = 8.03TPEE364 pKa = 3.85SCYY367 pKa = 10.65KK368 pKa = 10.23NKK370 pKa = 10.25SVYY373 pKa = 10.39DD374 pKa = 4.03LQPLLARR381 pKa = 11.84LDD383 pKa = 3.63IIEE386 pKa = 4.09IQKK389 pKa = 10.85FGDD392 pKa = 3.29IQVICTPPIEE402 pKa = 4.1EE403 pKa = 3.96VMMSEE408 pKa = 4.69FEE410 pKa = 4.64PIGITEE416 pKa = 5.12LYY418 pKa = 10.42SDD420 pKa = 4.04EE421 pKa = 4.82NGNIHH426 pKa = 7.04
MM1 pKa = 7.49NSVCTLSHH9 pKa = 7.12RR10 pKa = 11.84STSLLSATTTVTDD23 pKa = 3.38PHH25 pKa = 5.76TTTTEE30 pKa = 3.8TTRR33 pKa = 11.84PDD35 pKa = 2.99IVIRR39 pKa = 11.84PDD41 pKa = 3.38TLISANRR48 pKa = 11.84NQDD51 pKa = 3.48DD52 pKa = 3.98EE53 pKa = 4.87TGLEE57 pKa = 4.63PIPDD61 pKa = 3.28KK62 pKa = 11.2NFRR65 pKa = 11.84LNCKK69 pKa = 9.83AVFLTYY75 pKa = 8.8PQCALEE81 pKa = 4.47PEE83 pKa = 4.85HH84 pKa = 7.2VLQTILACNKK94 pKa = 8.17TKK96 pKa = 10.21HH97 pKa = 6.28CSVIVAQEE105 pKa = 3.7HH106 pKa = 6.38HH107 pKa = 6.91KK108 pKa = 11.07DD109 pKa = 3.8GNKK112 pKa = 9.62HH113 pKa = 4.28LHH115 pKa = 6.27VYY117 pKa = 10.92LEE119 pKa = 4.23FPKK122 pKa = 10.8KK123 pKa = 9.92IDD125 pKa = 3.17IKK127 pKa = 7.71TARR130 pKa = 11.84YY131 pKa = 8.76FNYY134 pKa = 8.73ITGKK138 pKa = 8.93QGNYY142 pKa = 7.83QKK144 pKa = 11.1VKK146 pKa = 10.6VRR148 pKa = 11.84GACIKK153 pKa = 9.32YY154 pKa = 8.2VCKK157 pKa = 10.09EE158 pKa = 3.72NRR160 pKa = 11.84YY161 pKa = 8.82VAHH164 pKa = 6.58QIDD167 pKa = 4.6VPTIIQNYY175 pKa = 7.86EE176 pKa = 3.58KK177 pKa = 10.46QQEE180 pKa = 4.39KK181 pKa = 10.49KK182 pKa = 10.42KK183 pKa = 10.87SRR185 pKa = 11.84NQAKK189 pKa = 9.95NKK191 pKa = 9.68RR192 pKa = 11.84VYY194 pKa = 10.74EE195 pKa = 4.21LIKK198 pKa = 10.39EE199 pKa = 4.29GKK201 pKa = 9.85KK202 pKa = 10.23YY203 pKa = 10.91EE204 pKa = 4.25EE205 pKa = 4.89LLLDD209 pKa = 4.23CTTGPYY215 pKa = 10.3CLLHH219 pKa = 6.12GNKK222 pKa = 8.76IKK224 pKa = 11.18NMITDD229 pKa = 4.16FEE231 pKa = 4.69EE232 pKa = 4.21IAMKK236 pKa = 9.79KK237 pKa = 8.6QRR239 pKa = 11.84IADD242 pKa = 4.11RR243 pKa = 11.84PAYY246 pKa = 10.12LHH248 pKa = 6.96LILPGKK254 pKa = 10.32DD255 pKa = 2.8IDD257 pKa = 4.75LLAKK261 pKa = 10.38KK262 pKa = 9.59PFKK265 pKa = 10.91SPQFWIFGPPNVGKK279 pKa = 8.97TSMLQSLFDD288 pKa = 4.12AGLKK292 pKa = 10.45GFEE295 pKa = 4.06IPINNDD301 pKa = 2.74FARR304 pKa = 11.84WDD306 pKa = 3.63DD307 pKa = 3.66EE308 pKa = 4.92LFDD311 pKa = 3.92FAYY314 pKa = 9.71IDD316 pKa = 3.69EE317 pKa = 5.06FKK319 pKa = 11.18GQLTIQFLNSFLQGSRR335 pKa = 11.84MALPGKK341 pKa = 9.61YY342 pKa = 10.31VIGGKK347 pKa = 9.3IKK349 pKa = 10.74NKK351 pKa = 10.21NIPTFILSNYY361 pKa = 8.03TPEE364 pKa = 3.85SCYY367 pKa = 10.65KK368 pKa = 10.23NKK370 pKa = 10.25SVYY373 pKa = 10.39DD374 pKa = 4.03LQPLLARR381 pKa = 11.84LDD383 pKa = 3.63IIEE386 pKa = 4.09IQKK389 pKa = 10.85FGDD392 pKa = 3.29IQVICTPPIEE402 pKa = 4.1EE403 pKa = 3.96VMMSEE408 pKa = 4.69FEE410 pKa = 4.64PIGITEE416 pKa = 5.12LYY418 pKa = 10.42SDD420 pKa = 4.04EE421 pKa = 4.82NGNIHH426 pKa = 7.04
Molecular weight: 48.83 kDa
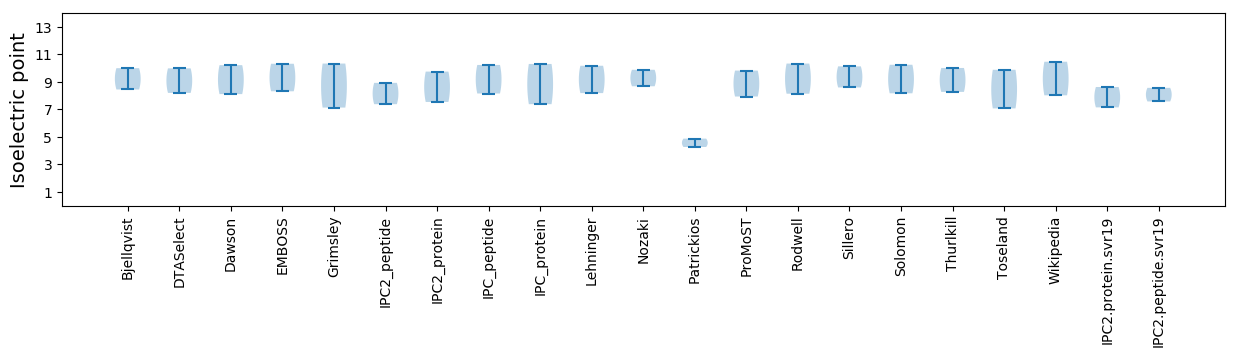
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A140CTW2|A0A140CTW2_9VIRU Replication-associated protein OS=Pacific flying fox faeces associated circular DNA virus-14 OX=1796009 PE=3 SV=1
MM1 pKa = 7.55NSNFNYY7 pKa = 10.12FLEE10 pKa = 4.33NYY12 pKa = 9.16KK13 pKa = 10.92LSGKK17 pKa = 9.73LLYY20 pKa = 10.25INPQLFQVFMMSGIPTNPNKK40 pKa = 9.82YY41 pKa = 9.69RR42 pKa = 11.84KK43 pKa = 9.17RR44 pKa = 11.84EE45 pKa = 3.75YY46 pKa = 10.01STISSRR52 pKa = 11.84NYY54 pKa = 10.56SNEE57 pKa = 3.62QEE59 pKa = 5.37DD60 pKa = 5.81DD61 pKa = 3.99YY62 pKa = 11.86QCLYY66 pKa = 11.12FNFRR70 pKa = 11.84RR71 pKa = 11.84QYY73 pKa = 9.27SKK75 pKa = 10.85YY76 pKa = 8.87RR77 pKa = 11.84NRR79 pKa = 11.84FPRR82 pKa = 11.84SYY84 pKa = 10.82GAYY87 pKa = 7.79TRR89 pKa = 11.84RR90 pKa = 11.84AVNVRR95 pKa = 11.84GLSYY99 pKa = 8.49PTGVRR104 pKa = 11.84GFWPGRR110 pKa = 11.84RR111 pKa = 11.84STFRR115 pKa = 11.84RR116 pKa = 11.84ITQNVRR122 pKa = 11.84TPSNRR127 pKa = 11.84IVRR130 pKa = 11.84MTYY133 pKa = 9.92SVPEE137 pKa = 3.95KK138 pKa = 10.79KK139 pKa = 10.54FIDD142 pKa = 3.67VTGTYY147 pKa = 10.71AVTAAGDD154 pKa = 3.73LTLLNGLTQGVSVISRR170 pKa = 11.84VGNKK174 pKa = 8.93IMMRR178 pKa = 11.84SLDD181 pKa = 3.82LRR183 pKa = 11.84WRR185 pKa = 11.84LTGAPFSATPTLPFNMVRR203 pKa = 11.84VMLVWDD209 pKa = 4.29AQPNGVIASAGDD221 pKa = 3.46ILQDD225 pKa = 3.55TTPGTGVVSPQQRR238 pKa = 11.84DD239 pKa = 3.44YY240 pKa = 10.68IQRR243 pKa = 11.84FRR245 pKa = 11.84IIMDD249 pKa = 3.74RR250 pKa = 11.84KK251 pKa = 8.65FTLTNLVANNGPTDD265 pKa = 4.05IIIACDD271 pKa = 3.36QMYY274 pKa = 10.76KK275 pKa = 8.89KK276 pKa = 8.7TKK278 pKa = 10.66LMVTYY283 pKa = 10.52SDD285 pKa = 4.46SNTGLIDD292 pKa = 5.36DD293 pKa = 4.78ISTGALYY300 pKa = 10.65LVTVGLHH307 pKa = 5.69AAAANQGVIEE317 pKa = 4.26YY318 pKa = 8.64FSRR321 pKa = 11.84VRR323 pKa = 11.84YY324 pKa = 9.45YY325 pKa = 11.42DD326 pKa = 3.18NN327 pKa = 4.15
MM1 pKa = 7.55NSNFNYY7 pKa = 10.12FLEE10 pKa = 4.33NYY12 pKa = 9.16KK13 pKa = 10.92LSGKK17 pKa = 9.73LLYY20 pKa = 10.25INPQLFQVFMMSGIPTNPNKK40 pKa = 9.82YY41 pKa = 9.69RR42 pKa = 11.84KK43 pKa = 9.17RR44 pKa = 11.84EE45 pKa = 3.75YY46 pKa = 10.01STISSRR52 pKa = 11.84NYY54 pKa = 10.56SNEE57 pKa = 3.62QEE59 pKa = 5.37DD60 pKa = 5.81DD61 pKa = 3.99YY62 pKa = 11.86QCLYY66 pKa = 11.12FNFRR70 pKa = 11.84RR71 pKa = 11.84QYY73 pKa = 9.27SKK75 pKa = 10.85YY76 pKa = 8.87RR77 pKa = 11.84NRR79 pKa = 11.84FPRR82 pKa = 11.84SYY84 pKa = 10.82GAYY87 pKa = 7.79TRR89 pKa = 11.84RR90 pKa = 11.84AVNVRR95 pKa = 11.84GLSYY99 pKa = 8.49PTGVRR104 pKa = 11.84GFWPGRR110 pKa = 11.84RR111 pKa = 11.84STFRR115 pKa = 11.84RR116 pKa = 11.84ITQNVRR122 pKa = 11.84TPSNRR127 pKa = 11.84IVRR130 pKa = 11.84MTYY133 pKa = 9.92SVPEE137 pKa = 3.95KK138 pKa = 10.79KK139 pKa = 10.54FIDD142 pKa = 3.67VTGTYY147 pKa = 10.71AVTAAGDD154 pKa = 3.73LTLLNGLTQGVSVISRR170 pKa = 11.84VGNKK174 pKa = 8.93IMMRR178 pKa = 11.84SLDD181 pKa = 3.82LRR183 pKa = 11.84WRR185 pKa = 11.84LTGAPFSATPTLPFNMVRR203 pKa = 11.84VMLVWDD209 pKa = 4.29AQPNGVIASAGDD221 pKa = 3.46ILQDD225 pKa = 3.55TTPGTGVVSPQQRR238 pKa = 11.84DD239 pKa = 3.44YY240 pKa = 10.68IQRR243 pKa = 11.84FRR245 pKa = 11.84IIMDD249 pKa = 3.74RR250 pKa = 11.84KK251 pKa = 8.65FTLTNLVANNGPTDD265 pKa = 4.05IIIACDD271 pKa = 3.36QMYY274 pKa = 10.76KK275 pKa = 8.89KK276 pKa = 8.7TKK278 pKa = 10.66LMVTYY283 pKa = 10.52SDD285 pKa = 4.46SNTGLIDD292 pKa = 5.36DD293 pKa = 4.78ISTGALYY300 pKa = 10.65LVTVGLHH307 pKa = 5.69AAAANQGVIEE317 pKa = 4.26YY318 pKa = 8.64FSRR321 pKa = 11.84VRR323 pKa = 11.84YY324 pKa = 9.45YY325 pKa = 11.42DD326 pKa = 3.18NN327 pKa = 4.15
Molecular weight: 37.43 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
753 |
327 |
426 |
376.5 |
43.13 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.781 ± 0.264 | 1.726 ± 0.703 |
5.312 ± 0.264 | 4.382 ± 1.607 |
4.515 ± 0.045 | 5.71 ± 0.642 |
1.726 ± 0.896 | 7.57 ± 1.11 |
7.039 ± 2.125 | 8.101 ± 0.48 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.39 ± 0.614 | 6.507 ± 0.525 |
5.179 ± 0.374 | 4.515 ± 0.148 |
5.976 ± 2.018 | 5.445 ± 1.002 |
7.437 ± 0.517 | 5.71 ± 1.028 |
0.664 ± 0.16 | 5.312 ± 0.893 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
