
Methanoculleus taiwanensis
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Methanomicrobia; Methanomicrobiales; Methanomicrobiaceae; Methanoculleus
Average proteome isoelectric point is 5.82
Get precalculated fractions of proteins
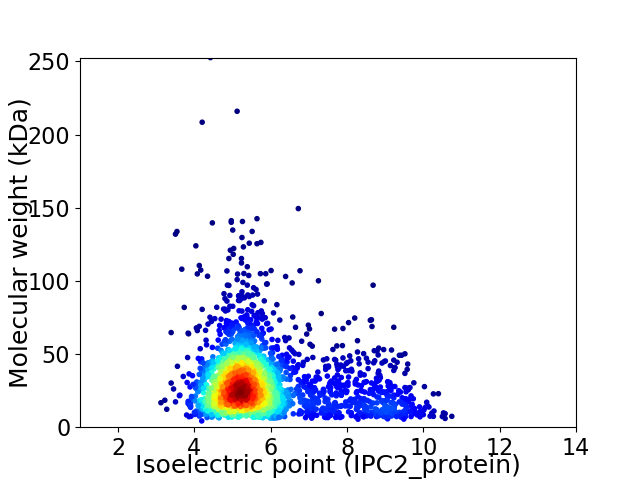
Virtual 2D-PAGE plot for 2433 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A498H2Q6|A0A498H2Q6_9EURY Acyl_transf_3 domain-containing protein OS=Methanoculleus taiwanensis OX=1550565 GN=ABH15_04610 PE=4 SV=1
MM1 pKa = 7.75RR2 pKa = 11.84KK3 pKa = 8.94ILLISLVILACIVTLLPSAAAASEE27 pKa = 4.33LPAGGQSSTDD37 pKa = 3.19VSVSTNTSVTSTTDD51 pKa = 3.11VSVTTGAGGGAPAPNGTRR69 pKa = 11.84AVDD72 pKa = 3.24VALAIDD78 pKa = 4.45SSASMRR84 pKa = 11.84NTDD87 pKa = 4.15PDD89 pKa = 3.97DD90 pKa = 4.1LRR92 pKa = 11.84IDD94 pKa = 3.7AARR97 pKa = 11.84LFVDD101 pKa = 5.22LAANDD106 pKa = 4.15DD107 pKa = 4.06LLAIVDD113 pKa = 4.52FDD115 pKa = 3.96HH116 pKa = 7.11AVTVLEE122 pKa = 4.16PLSLVRR128 pKa = 11.84NNRR131 pKa = 11.84TALKK135 pKa = 9.51MAIDD139 pKa = 3.98QVDD142 pKa = 3.3SSGEE146 pKa = 4.09TNIGGGLLASFDD158 pKa = 3.82QLAEE162 pKa = 4.07RR163 pKa = 11.84GIPGHH168 pKa = 6.37GKK170 pKa = 8.51ATVLLTDD177 pKa = 4.49GVHH180 pKa = 5.64NTGTDD185 pKa = 3.5PLAVVPLYY193 pKa = 10.87VEE195 pKa = 4.26QGWPVYY201 pKa = 9.47TVALTDD207 pKa = 4.0FADD210 pKa = 3.8EE211 pKa = 3.91ALMRR215 pKa = 11.84EE216 pKa = 4.63IARR219 pKa = 11.84QTGGRR224 pKa = 11.84YY225 pKa = 9.14YY226 pKa = 10.87SAPTSSALLDD236 pKa = 3.16IYY238 pKa = 11.42NRR240 pKa = 11.84IKK242 pKa = 9.75QTIKK246 pKa = 10.79GQDD249 pKa = 3.3QITSEE254 pKa = 4.16TGTISVNQTVQNIVNIDD271 pKa = 3.34TSVQVVEE278 pKa = 5.61FTCLCPQTTINYY290 pKa = 8.77YY291 pKa = 9.68IYY293 pKa = 9.89YY294 pKa = 9.38PNGTRR299 pKa = 11.84VEE301 pKa = 4.59LDD303 pKa = 3.44PEE305 pKa = 4.66SPTGTEE311 pKa = 4.42SPDD314 pKa = 2.81ITYY317 pKa = 10.18ISSDD321 pKa = 3.28TYY323 pKa = 11.09QIYY326 pKa = 9.57QVNNPVVGTWIEE338 pKa = 4.3VINATDD344 pKa = 3.46ANVTVDD350 pKa = 3.4YY351 pKa = 10.51SSTVAVSATIEE362 pKa = 3.69LAAAIDD368 pKa = 3.92AFSYY372 pKa = 11.18APGEE376 pKa = 4.23PVTVTAEE383 pKa = 4.12VRR385 pKa = 11.84NEE387 pKa = 3.64TAGIEE392 pKa = 4.28NATVTAVFTLPGGEE406 pKa = 4.19NEE408 pKa = 4.37TVLLNGTGNGTYY420 pKa = 10.56AGTFSNTSTTGAYY433 pKa = 8.76QVQVRR438 pKa = 11.84STVGDD443 pKa = 3.38IVRR446 pKa = 11.84QEE448 pKa = 4.13VLDD451 pKa = 3.94FAVTEE456 pKa = 4.11EE457 pKa = 4.65APPVTPTPTMAPNATPATPNATPPPTMTLNEE488 pKa = 4.54TPTMTLNEE496 pKa = 4.72TPTMTPTPTPTVPVEE511 pKa = 4.17QPSGAFLQVTNNSADD526 pKa = 3.37QVNPDD531 pKa = 2.88ISGDD535 pKa = 3.78LVVWQDD541 pKa = 3.26NRR543 pKa = 11.84SGNSDD548 pKa = 2.86IFRR551 pKa = 11.84YY552 pKa = 10.26NLTDD556 pKa = 3.61GAVTQITTDD565 pKa = 3.64PADD568 pKa = 3.42QANPAVSGVSRR579 pKa = 11.84LIVWEE584 pKa = 4.22DD585 pKa = 3.24NRR587 pKa = 11.84NGDD590 pKa = 3.68WDD592 pKa = 3.94IFWYY596 pKa = 10.66SPFSGEE602 pKa = 4.12EE603 pKa = 4.17FPVTTAPGDD612 pKa = 3.35QRR614 pKa = 11.84YY615 pKa = 8.67PSIAFQYY622 pKa = 10.42VCWQDD627 pKa = 3.13NRR629 pKa = 11.84NGTWTAYY636 pKa = 10.16VADD639 pKa = 6.03LIPGQEE645 pKa = 4.1TGSPAGMPGSVQEE658 pKa = 4.34NPAIAFGGVDD668 pKa = 5.43GPTVWQDD675 pKa = 3.23DD676 pKa = 4.13RR677 pKa = 11.84NGSFDD682 pKa = 4.18IYY684 pKa = 10.27WSNAGTNEE692 pKa = 3.94MPVCIDD698 pKa = 4.28AGDD701 pKa = 3.76QQFPAIDD708 pKa = 3.87GLSVVWQDD716 pKa = 3.29TRR718 pKa = 11.84NGSSDD723 pKa = 3.02IYY725 pKa = 11.12LGDD728 pKa = 4.16LEE730 pKa = 4.79TGNRR734 pKa = 11.84TQITDD739 pKa = 3.48DD740 pKa = 4.47PADD743 pKa = 3.36QVYY746 pKa = 10.48PDD748 pKa = 3.74ISGDD752 pKa = 3.76FIVWQDD758 pKa = 3.36NRR760 pKa = 11.84NGNWDD765 pKa = 3.06IFLYY769 pKa = 10.79NRR771 pKa = 11.84VDD773 pKa = 3.36GTLVQVTDD781 pKa = 4.54DD782 pKa = 4.67PADD785 pKa = 3.42QIYY788 pKa = 9.96PSIAGNYY795 pKa = 9.11IVWQDD800 pKa = 3.33NRR802 pKa = 11.84SGNWDD807 pKa = 3.21IFLYY811 pKa = 10.74AIDD814 pKa = 4.51GVLPASPAEE823 pKa = 4.1TGGMPEE829 pKa = 4.64PGLPPLAANLTANVTNGTAPLTVLFNATTSGDD861 pKa = 3.44PTAWSWEE868 pKa = 4.17FGDD871 pKa = 4.31GATATGEE878 pKa = 4.1GSVQYY883 pKa = 9.81TYY885 pKa = 11.04QSPGTYY891 pKa = 7.74TARR894 pKa = 11.84LTVTNEE900 pKa = 3.72TGGNATDD907 pKa = 3.8TVIVTVTQEE916 pKa = 3.68PAEE919 pKa = 4.16NVTDD923 pKa = 3.8NQTPEE928 pKa = 3.56AALRR932 pKa = 11.84ADD934 pKa = 4.19PEE936 pKa = 4.43TGAVPLTVTFQDD948 pKa = 3.37QSTGSIDD955 pKa = 2.77EE956 pKa = 4.3WLWFFGDD963 pKa = 4.12GNASASPNATHH974 pKa = 7.16TYY976 pKa = 10.3VEE978 pKa = 4.36AGNYY982 pKa = 7.36TVTLTVSGPAGSDD995 pKa = 3.25TANGTIAVLPAIPAPEE1011 pKa = 3.79EE1012 pKa = 3.78AAGNLARR1019 pKa = 11.84AA1020 pKa = 4.07
MM1 pKa = 7.75RR2 pKa = 11.84KK3 pKa = 8.94ILLISLVILACIVTLLPSAAAASEE27 pKa = 4.33LPAGGQSSTDD37 pKa = 3.19VSVSTNTSVTSTTDD51 pKa = 3.11VSVTTGAGGGAPAPNGTRR69 pKa = 11.84AVDD72 pKa = 3.24VALAIDD78 pKa = 4.45SSASMRR84 pKa = 11.84NTDD87 pKa = 4.15PDD89 pKa = 3.97DD90 pKa = 4.1LRR92 pKa = 11.84IDD94 pKa = 3.7AARR97 pKa = 11.84LFVDD101 pKa = 5.22LAANDD106 pKa = 4.15DD107 pKa = 4.06LLAIVDD113 pKa = 4.52FDD115 pKa = 3.96HH116 pKa = 7.11AVTVLEE122 pKa = 4.16PLSLVRR128 pKa = 11.84NNRR131 pKa = 11.84TALKK135 pKa = 9.51MAIDD139 pKa = 3.98QVDD142 pKa = 3.3SSGEE146 pKa = 4.09TNIGGGLLASFDD158 pKa = 3.82QLAEE162 pKa = 4.07RR163 pKa = 11.84GIPGHH168 pKa = 6.37GKK170 pKa = 8.51ATVLLTDD177 pKa = 4.49GVHH180 pKa = 5.64NTGTDD185 pKa = 3.5PLAVVPLYY193 pKa = 10.87VEE195 pKa = 4.26QGWPVYY201 pKa = 9.47TVALTDD207 pKa = 4.0FADD210 pKa = 3.8EE211 pKa = 3.91ALMRR215 pKa = 11.84EE216 pKa = 4.63IARR219 pKa = 11.84QTGGRR224 pKa = 11.84YY225 pKa = 9.14YY226 pKa = 10.87SAPTSSALLDD236 pKa = 3.16IYY238 pKa = 11.42NRR240 pKa = 11.84IKK242 pKa = 9.75QTIKK246 pKa = 10.79GQDD249 pKa = 3.3QITSEE254 pKa = 4.16TGTISVNQTVQNIVNIDD271 pKa = 3.34TSVQVVEE278 pKa = 5.61FTCLCPQTTINYY290 pKa = 8.77YY291 pKa = 9.68IYY293 pKa = 9.89YY294 pKa = 9.38PNGTRR299 pKa = 11.84VEE301 pKa = 4.59LDD303 pKa = 3.44PEE305 pKa = 4.66SPTGTEE311 pKa = 4.42SPDD314 pKa = 2.81ITYY317 pKa = 10.18ISSDD321 pKa = 3.28TYY323 pKa = 11.09QIYY326 pKa = 9.57QVNNPVVGTWIEE338 pKa = 4.3VINATDD344 pKa = 3.46ANVTVDD350 pKa = 3.4YY351 pKa = 10.51SSTVAVSATIEE362 pKa = 3.69LAAAIDD368 pKa = 3.92AFSYY372 pKa = 11.18APGEE376 pKa = 4.23PVTVTAEE383 pKa = 4.12VRR385 pKa = 11.84NEE387 pKa = 3.64TAGIEE392 pKa = 4.28NATVTAVFTLPGGEE406 pKa = 4.19NEE408 pKa = 4.37TVLLNGTGNGTYY420 pKa = 10.56AGTFSNTSTTGAYY433 pKa = 8.76QVQVRR438 pKa = 11.84STVGDD443 pKa = 3.38IVRR446 pKa = 11.84QEE448 pKa = 4.13VLDD451 pKa = 3.94FAVTEE456 pKa = 4.11EE457 pKa = 4.65APPVTPTPTMAPNATPATPNATPPPTMTLNEE488 pKa = 4.54TPTMTLNEE496 pKa = 4.72TPTMTPTPTPTVPVEE511 pKa = 4.17QPSGAFLQVTNNSADD526 pKa = 3.37QVNPDD531 pKa = 2.88ISGDD535 pKa = 3.78LVVWQDD541 pKa = 3.26NRR543 pKa = 11.84SGNSDD548 pKa = 2.86IFRR551 pKa = 11.84YY552 pKa = 10.26NLTDD556 pKa = 3.61GAVTQITTDD565 pKa = 3.64PADD568 pKa = 3.42QANPAVSGVSRR579 pKa = 11.84LIVWEE584 pKa = 4.22DD585 pKa = 3.24NRR587 pKa = 11.84NGDD590 pKa = 3.68WDD592 pKa = 3.94IFWYY596 pKa = 10.66SPFSGEE602 pKa = 4.12EE603 pKa = 4.17FPVTTAPGDD612 pKa = 3.35QRR614 pKa = 11.84YY615 pKa = 8.67PSIAFQYY622 pKa = 10.42VCWQDD627 pKa = 3.13NRR629 pKa = 11.84NGTWTAYY636 pKa = 10.16VADD639 pKa = 6.03LIPGQEE645 pKa = 4.1TGSPAGMPGSVQEE658 pKa = 4.34NPAIAFGGVDD668 pKa = 5.43GPTVWQDD675 pKa = 3.23DD676 pKa = 4.13RR677 pKa = 11.84NGSFDD682 pKa = 4.18IYY684 pKa = 10.27WSNAGTNEE692 pKa = 3.94MPVCIDD698 pKa = 4.28AGDD701 pKa = 3.76QQFPAIDD708 pKa = 3.87GLSVVWQDD716 pKa = 3.29TRR718 pKa = 11.84NGSSDD723 pKa = 3.02IYY725 pKa = 11.12LGDD728 pKa = 4.16LEE730 pKa = 4.79TGNRR734 pKa = 11.84TQITDD739 pKa = 3.48DD740 pKa = 4.47PADD743 pKa = 3.36QVYY746 pKa = 10.48PDD748 pKa = 3.74ISGDD752 pKa = 3.76FIVWQDD758 pKa = 3.36NRR760 pKa = 11.84NGNWDD765 pKa = 3.06IFLYY769 pKa = 10.79NRR771 pKa = 11.84VDD773 pKa = 3.36GTLVQVTDD781 pKa = 4.54DD782 pKa = 4.67PADD785 pKa = 3.42QIYY788 pKa = 9.96PSIAGNYY795 pKa = 9.11IVWQDD800 pKa = 3.33NRR802 pKa = 11.84SGNWDD807 pKa = 3.21IFLYY811 pKa = 10.74AIDD814 pKa = 4.51GVLPASPAEE823 pKa = 4.1TGGMPEE829 pKa = 4.64PGLPPLAANLTANVTNGTAPLTVLFNATTSGDD861 pKa = 3.44PTAWSWEE868 pKa = 4.17FGDD871 pKa = 4.31GATATGEE878 pKa = 4.1GSVQYY883 pKa = 9.81TYY885 pKa = 11.04QSPGTYY891 pKa = 7.74TARR894 pKa = 11.84LTVTNEE900 pKa = 3.72TGGNATDD907 pKa = 3.8TVIVTVTQEE916 pKa = 3.68PAEE919 pKa = 4.16NVTDD923 pKa = 3.8NQTPEE928 pKa = 3.56AALRR932 pKa = 11.84ADD934 pKa = 4.19PEE936 pKa = 4.43TGAVPLTVTFQDD948 pKa = 3.37QSTGSIDD955 pKa = 2.77EE956 pKa = 4.3WLWFFGDD963 pKa = 4.12GNASASPNATHH974 pKa = 7.16TYY976 pKa = 10.3VEE978 pKa = 4.36AGNYY982 pKa = 7.36TVTLTVSGPAGSDD995 pKa = 3.25TANGTIAVLPAIPAPEE1011 pKa = 3.79EE1012 pKa = 3.78AAGNLARR1019 pKa = 11.84AA1020 pKa = 4.07
Molecular weight: 107.99 kDa
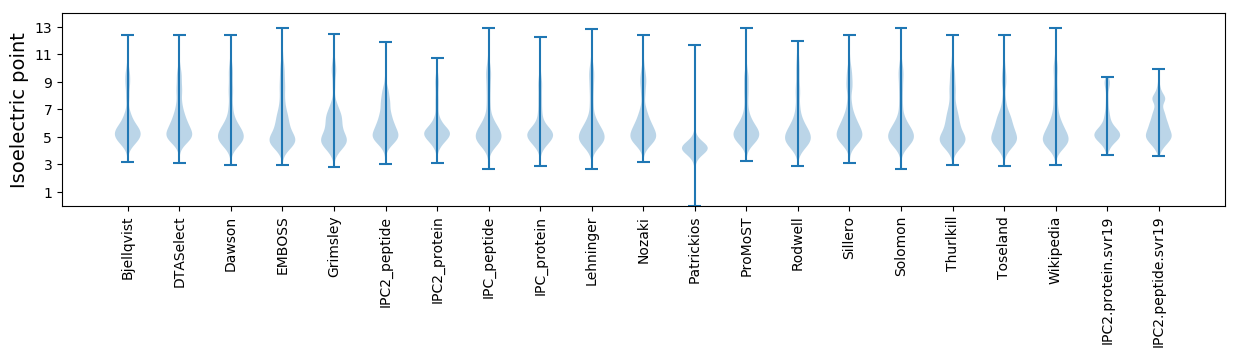
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A498GWT7|A0A498GWT7_9EURY Uncharacterized protein OS=Methanoculleus taiwanensis OX=1550565 GN=ABH15_12600 PE=4 SV=1
MM1 pKa = 7.68PAKK4 pKa = 9.89QGRR7 pKa = 11.84DD8 pKa = 2.86IVQLRR13 pKa = 11.84FRR15 pKa = 11.84DD16 pKa = 3.82PGIAAPLTPARR27 pKa = 11.84PDD29 pKa = 3.43DD30 pKa = 3.89VPEE33 pKa = 4.23RR34 pKa = 11.84NAMPFEE40 pKa = 4.23YY41 pKa = 10.83GNLSGVWKK49 pKa = 10.28IAGIPKK55 pKa = 9.76RR56 pKa = 11.84SAIMRR61 pKa = 11.84QNWFCRR67 pKa = 11.84CRR69 pKa = 3.39
MM1 pKa = 7.68PAKK4 pKa = 9.89QGRR7 pKa = 11.84DD8 pKa = 2.86IVQLRR13 pKa = 11.84FRR15 pKa = 11.84DD16 pKa = 3.82PGIAAPLTPARR27 pKa = 11.84PDD29 pKa = 3.43DD30 pKa = 3.89VPEE33 pKa = 4.23RR34 pKa = 11.84NAMPFEE40 pKa = 4.23YY41 pKa = 10.83GNLSGVWKK49 pKa = 10.28IAGIPKK55 pKa = 9.76RR56 pKa = 11.84SAIMRR61 pKa = 11.84QNWFCRR67 pKa = 11.84CRR69 pKa = 3.39
Molecular weight: 7.87 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
704774 |
41 |
2405 |
289.7 |
31.76 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.955 ± 0.064 | 1.298 ± 0.023 |
5.631 ± 0.04 | 7.005 ± 0.054 |
3.651 ± 0.036 | 8.428 ± 0.056 |
1.945 ± 0.022 | 6.684 ± 0.042 |
3.444 ± 0.047 | 9.575 ± 0.059 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.519 ± 0.026 | 2.715 ± 0.034 |
4.729 ± 0.034 | 2.537 ± 0.025 |
6.716 ± 0.054 | 5.332 ± 0.031 |
5.78 ± 0.046 | 8.015 ± 0.043 |
0.985 ± 0.02 | 3.058 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
