
Streptomyces phage Lannister
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; Arquatrovirinae; Likavirus; Streptomyces virus Lannister
Average proteome isoelectric point is 6.05
Get precalculated fractions of proteins
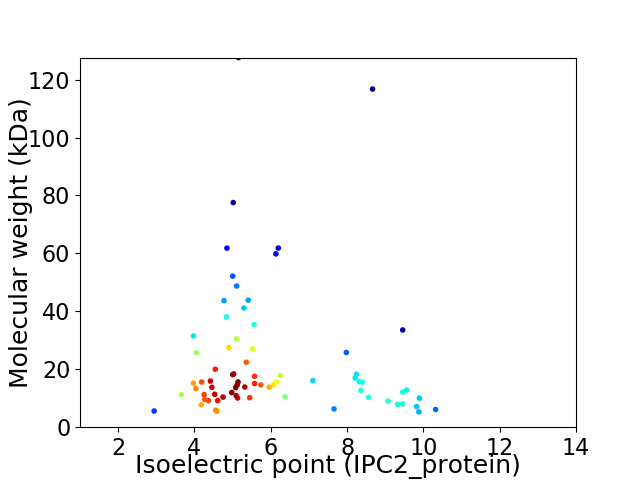
Virtual 2D-PAGE plot for 73 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
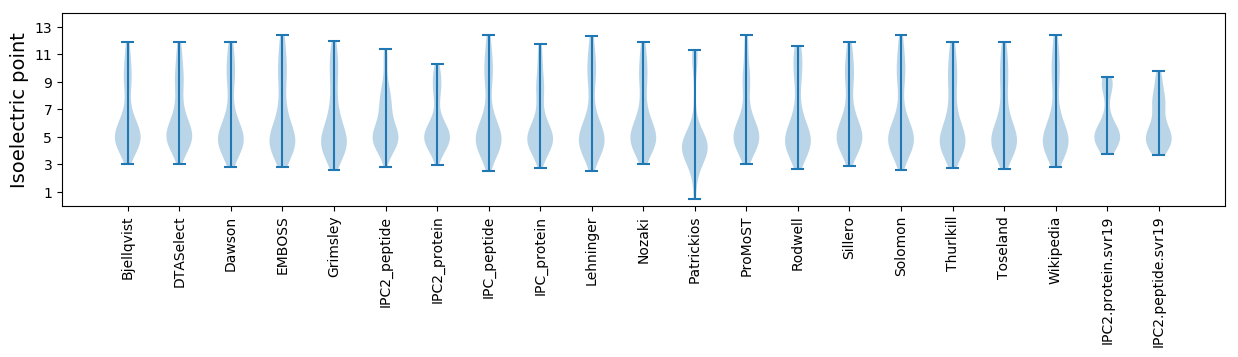
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0K1Y9V9|A0A0K1Y9V9_9CAUD Uncharacterized protein OS=Streptomyces phage Lannister OX=1674927 GN=SEA_LANNISTER_1 PE=4 SV=1
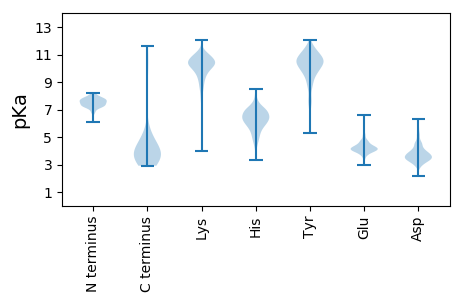
MM1 pKa = 7.22NVDD4 pKa = 2.8IDD6 pKa = 4.35TVQEE10 pKa = 4.09YY11 pKa = 11.2ADD13 pKa = 3.75NALEE17 pKa = 4.36RR18 pKa = 11.84PSDD21 pKa = 3.4AAFWDD26 pKa = 3.92SRR28 pKa = 11.84CYY30 pKa = 7.57EE31 pKa = 3.89THH33 pKa = 5.99VPVIGWADD41 pKa = 3.62RR42 pKa = 11.84GDD44 pKa = 4.95DD45 pKa = 3.38ILEE48 pKa = 4.06EE49 pKa = 4.48SNYY52 pKa = 10.26HH53 pKa = 4.92SAKK56 pKa = 10.63SLIEE60 pKa = 4.15GAATDD65 pKa = 4.12PEE67 pKa = 4.7HH68 pKa = 7.97VFEE71 pKa = 5.15GSAGHH76 pKa = 6.45WLVGSLSQVWVQVYY90 pKa = 7.04EE91 pKa = 4.16TRR93 pKa = 11.84PEE95 pKa = 4.19CDD97 pKa = 2.83TIGCEE102 pKa = 4.11EE103 pKa = 4.49EE104 pKa = 4.01ATEE107 pKa = 4.19VASYY111 pKa = 10.11TRR113 pKa = 11.84TSGEE117 pKa = 3.78GHH119 pKa = 6.7FCRR122 pKa = 11.84DD123 pKa = 2.92HH124 pKa = 7.18ALPLLYY130 pKa = 10.7SPIGEE135 pKa = 4.19LLGVEE140 pKa = 5.14LEE142 pKa = 4.1DD143 pKa = 4.73LPRR146 pKa = 11.84EE147 pKa = 3.9FTAAFIEE154 pKa = 4.3AVEE157 pKa = 4.07IQEE160 pKa = 4.21ALKK163 pKa = 10.44DD164 pKa = 3.87YY165 pKa = 10.57PIVDD169 pKa = 3.39EE170 pKa = 4.73SDD172 pKa = 3.15FSEE175 pKa = 4.7RR176 pKa = 11.84EE177 pKa = 3.71WVRR180 pKa = 11.84FEE182 pKa = 4.51EE183 pKa = 4.26NCKK186 pKa = 10.07EE187 pKa = 4.15ALEE190 pKa = 4.19QAQRR194 pKa = 11.84EE195 pKa = 4.29YY196 pKa = 11.21EE197 pKa = 4.15DD198 pKa = 5.73DD199 pKa = 3.83SVEE202 pKa = 4.74DD203 pKa = 3.39EE204 pKa = 4.46TEE206 pKa = 3.51IEE208 pKa = 3.83NRR210 pKa = 11.84IFSEE214 pKa = 4.31SALSDD219 pKa = 3.54LFGYY223 pKa = 7.72EE224 pKa = 3.92ANAEE228 pKa = 4.24VSWEE232 pKa = 4.2SVAEE236 pKa = 4.27IYY238 pKa = 10.84AEE240 pKa = 3.9YY241 pKa = 10.36RR242 pKa = 11.84DD243 pKa = 4.18AYY245 pKa = 9.72FLEE248 pKa = 4.93RR249 pKa = 11.84ATEE252 pKa = 4.16VYY254 pKa = 9.45RR255 pKa = 11.84WNVLGYY261 pKa = 10.83NPDD264 pKa = 3.47QLEE267 pKa = 4.22LDD269 pKa = 3.4IVIIVAAA276 pKa = 4.47
MM1 pKa = 7.22NVDD4 pKa = 2.8IDD6 pKa = 4.35TVQEE10 pKa = 4.09YY11 pKa = 11.2ADD13 pKa = 3.75NALEE17 pKa = 4.36RR18 pKa = 11.84PSDD21 pKa = 3.4AAFWDD26 pKa = 3.92SRR28 pKa = 11.84CYY30 pKa = 7.57EE31 pKa = 3.89THH33 pKa = 5.99VPVIGWADD41 pKa = 3.62RR42 pKa = 11.84GDD44 pKa = 4.95DD45 pKa = 3.38ILEE48 pKa = 4.06EE49 pKa = 4.48SNYY52 pKa = 10.26HH53 pKa = 4.92SAKK56 pKa = 10.63SLIEE60 pKa = 4.15GAATDD65 pKa = 4.12PEE67 pKa = 4.7HH68 pKa = 7.97VFEE71 pKa = 5.15GSAGHH76 pKa = 6.45WLVGSLSQVWVQVYY90 pKa = 7.04EE91 pKa = 4.16TRR93 pKa = 11.84PEE95 pKa = 4.19CDD97 pKa = 2.83TIGCEE102 pKa = 4.11EE103 pKa = 4.49EE104 pKa = 4.01ATEE107 pKa = 4.19VASYY111 pKa = 10.11TRR113 pKa = 11.84TSGEE117 pKa = 3.78GHH119 pKa = 6.7FCRR122 pKa = 11.84DD123 pKa = 2.92HH124 pKa = 7.18ALPLLYY130 pKa = 10.7SPIGEE135 pKa = 4.19LLGVEE140 pKa = 5.14LEE142 pKa = 4.1DD143 pKa = 4.73LPRR146 pKa = 11.84EE147 pKa = 3.9FTAAFIEE154 pKa = 4.3AVEE157 pKa = 4.07IQEE160 pKa = 4.21ALKK163 pKa = 10.44DD164 pKa = 3.87YY165 pKa = 10.57PIVDD169 pKa = 3.39EE170 pKa = 4.73SDD172 pKa = 3.15FSEE175 pKa = 4.7RR176 pKa = 11.84EE177 pKa = 3.71WVRR180 pKa = 11.84FEE182 pKa = 4.51EE183 pKa = 4.26NCKK186 pKa = 10.07EE187 pKa = 4.15ALEE190 pKa = 4.19QAQRR194 pKa = 11.84EE195 pKa = 4.29YY196 pKa = 11.21EE197 pKa = 4.15DD198 pKa = 5.73DD199 pKa = 3.83SVEE202 pKa = 4.74DD203 pKa = 3.39EE204 pKa = 4.46TEE206 pKa = 3.51IEE208 pKa = 3.83NRR210 pKa = 11.84IFSEE214 pKa = 4.31SALSDD219 pKa = 3.54LFGYY223 pKa = 7.72EE224 pKa = 3.92ANAEE228 pKa = 4.24VSWEE232 pKa = 4.2SVAEE236 pKa = 4.27IYY238 pKa = 10.84AEE240 pKa = 3.9YY241 pKa = 10.36RR242 pKa = 11.84DD243 pKa = 4.18AYY245 pKa = 9.72FLEE248 pKa = 4.93RR249 pKa = 11.84ATEE252 pKa = 4.16VYY254 pKa = 9.45RR255 pKa = 11.84WNVLGYY261 pKa = 10.83NPDD264 pKa = 3.47QLEE267 pKa = 4.22LDD269 pKa = 3.4IVIIVAAA276 pKa = 4.47
Molecular weight: 31.49 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0K1Y9B5|A0A0K1Y9B5_9CAUD Tail assembly chaperone OS=Streptomyces phage Lannister OX=1674927 GN=SEA_LANNISTER_18 PE=4 SV=1
MM1 pKa = 7.22SVQRR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84GQVARR12 pKa = 11.84IWKK15 pKa = 4.65TTKK18 pKa = 10.72VIDD21 pKa = 3.44NRR23 pKa = 11.84GNEE26 pKa = 3.77VHH28 pKa = 6.31VAHH31 pKa = 7.12NDD33 pKa = 3.45GPHH36 pKa = 4.82VVRR39 pKa = 11.84AAFIPQRR46 pKa = 11.84SAKK49 pKa = 10.63AEE51 pKa = 4.13VPGQMQINITRR62 pKa = 11.84MIVAADD68 pKa = 3.77LEE70 pKa = 4.88GVEE73 pKa = 4.06LWSRR77 pKa = 11.84VEE79 pKa = 4.01WNGKK83 pKa = 6.9QWDD86 pKa = 4.06IVTPPAYY93 pKa = 10.01HH94 pKa = 6.38HH95 pKa = 6.93GSRR98 pKa = 11.84QTRR101 pKa = 11.84HH102 pKa = 4.11WSIDD106 pKa = 2.86IRR108 pKa = 11.84EE109 pKa = 4.09RR110 pKa = 11.84PP111 pKa = 3.36
MM1 pKa = 7.22SVQRR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84GQVARR12 pKa = 11.84IWKK15 pKa = 4.65TTKK18 pKa = 10.72VIDD21 pKa = 3.44NRR23 pKa = 11.84GNEE26 pKa = 3.77VHH28 pKa = 6.31VAHH31 pKa = 7.12NDD33 pKa = 3.45GPHH36 pKa = 4.82VVRR39 pKa = 11.84AAFIPQRR46 pKa = 11.84SAKK49 pKa = 10.63AEE51 pKa = 4.13VPGQMQINITRR62 pKa = 11.84MIVAADD68 pKa = 3.77LEE70 pKa = 4.88GVEE73 pKa = 4.06LWSRR77 pKa = 11.84VEE79 pKa = 4.01WNGKK83 pKa = 6.9QWDD86 pKa = 4.06IVTPPAYY93 pKa = 10.01HH94 pKa = 6.38HH95 pKa = 6.93GSRR98 pKa = 11.84QTRR101 pKa = 11.84HH102 pKa = 4.11WSIDD106 pKa = 2.86IRR108 pKa = 11.84EE109 pKa = 4.09RR110 pKa = 11.84PP111 pKa = 3.36
Molecular weight: 12.83 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
15216 |
43 |
1159 |
208.4 |
22.88 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.555 ± 0.3 | 0.69 ± 0.117 |
6.342 ± 0.225 | 7.249 ± 0.483 |
3.273 ± 0.165 | 8.465 ± 0.391 |
2.018 ± 0.194 | 4.397 ± 0.334 |
4.403 ± 0.276 | 8.498 ± 0.383 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.064 ± 0.139 | 3.023 ± 0.137 |
4.896 ± 0.219 | 3.293 ± 0.147 |
6.467 ± 0.368 | 5.948 ± 0.256 |
6.513 ± 0.358 | 7.302 ± 0.235 |
1.88 ± 0.145 | 2.727 ± 0.235 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
