
Radi vesiculovirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Alpharhabdovirinae; Vesiculovirus
Average proteome isoelectric point is 6.56
Get precalculated fractions of proteins
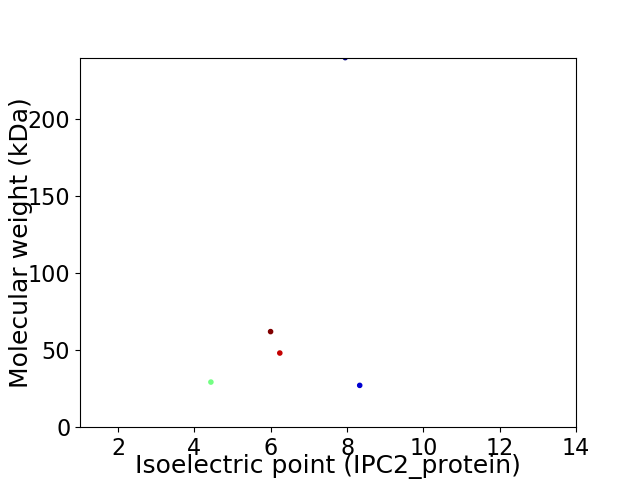
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0D3R1W3|A0A0D3R1W3_9RHAB Glycoprotein OS=Radi vesiculovirus OX=1972566 PE=3 SV=1
MM1 pKa = 8.08DD2 pKa = 4.74LQRR5 pKa = 11.84SAFEE9 pKa = 4.05ALRR12 pKa = 11.84NYY14 pKa = 9.87PNLEE18 pKa = 3.93NALADD23 pKa = 3.91ADD25 pKa = 4.0EE26 pKa = 4.91SEE28 pKa = 4.64SLRR31 pKa = 11.84DD32 pKa = 3.57PPSPNSSQAHH42 pKa = 4.98EE43 pKa = 4.35SEE45 pKa = 4.41GEE47 pKa = 3.99KK48 pKa = 10.52FYY50 pKa = 10.64LTDD53 pKa = 3.69QLDD56 pKa = 3.89LLEE59 pKa = 5.38SSDD62 pKa = 5.26EE63 pKa = 4.04EE64 pKa = 5.69DD65 pKa = 3.85EE66 pKa = 5.63DD67 pKa = 4.79LSTDD71 pKa = 3.11IGSSGQHH78 pKa = 5.45QDD80 pKa = 2.76TYY82 pKa = 11.9SNIDD86 pKa = 3.7DD87 pKa = 4.19RR88 pKa = 11.84SASNQDD94 pKa = 2.84EE95 pKa = 4.31DD96 pKa = 4.64SEE98 pKa = 4.67EE99 pKa = 3.77EE100 pKa = 3.94RR101 pKa = 11.84EE102 pKa = 4.25AYY104 pKa = 9.6GVHH107 pKa = 6.36FPSEE111 pKa = 4.36KK112 pKa = 9.28PTIDD116 pKa = 5.23LSDD119 pKa = 4.22LPATLQRR126 pKa = 11.84RR127 pKa = 11.84IFVDD131 pKa = 3.49ISNAFKK137 pKa = 11.27KK138 pKa = 10.28MMEE141 pKa = 4.44APSCQHH147 pKa = 5.7FPVMVRR153 pKa = 11.84GRR155 pKa = 11.84IEE157 pKa = 5.33LIPSSDD163 pKa = 3.39MAIHH167 pKa = 5.26QTPSSATTPEE177 pKa = 4.43SEE179 pKa = 5.14DD180 pKa = 3.99LPPSPVKK187 pKa = 10.12RR188 pKa = 11.84DD189 pKa = 3.33RR190 pKa = 11.84LLDD193 pKa = 3.35RR194 pKa = 11.84FKK196 pKa = 10.27IHH198 pKa = 6.7HH199 pKa = 7.21RR200 pKa = 11.84STQDD204 pKa = 3.31PLEE207 pKa = 4.23TSFVEE212 pKa = 4.13IFGSEE217 pKa = 3.94DD218 pKa = 2.96SARR221 pKa = 11.84AAWGDD226 pKa = 3.26GSLSYY231 pKa = 10.97KK232 pKa = 10.29EE233 pKa = 4.65LLILGLRR240 pKa = 11.84RR241 pKa = 11.84KK242 pKa = 10.1GVFNRR247 pKa = 11.84MRR249 pKa = 11.84IAYY252 pKa = 8.71DD253 pKa = 3.25LTPIFEE259 pKa = 4.36
MM1 pKa = 8.08DD2 pKa = 4.74LQRR5 pKa = 11.84SAFEE9 pKa = 4.05ALRR12 pKa = 11.84NYY14 pKa = 9.87PNLEE18 pKa = 3.93NALADD23 pKa = 3.91ADD25 pKa = 4.0EE26 pKa = 4.91SEE28 pKa = 4.64SLRR31 pKa = 11.84DD32 pKa = 3.57PPSPNSSQAHH42 pKa = 4.98EE43 pKa = 4.35SEE45 pKa = 4.41GEE47 pKa = 3.99KK48 pKa = 10.52FYY50 pKa = 10.64LTDD53 pKa = 3.69QLDD56 pKa = 3.89LLEE59 pKa = 5.38SSDD62 pKa = 5.26EE63 pKa = 4.04EE64 pKa = 5.69DD65 pKa = 3.85EE66 pKa = 5.63DD67 pKa = 4.79LSTDD71 pKa = 3.11IGSSGQHH78 pKa = 5.45QDD80 pKa = 2.76TYY82 pKa = 11.9SNIDD86 pKa = 3.7DD87 pKa = 4.19RR88 pKa = 11.84SASNQDD94 pKa = 2.84EE95 pKa = 4.31DD96 pKa = 4.64SEE98 pKa = 4.67EE99 pKa = 3.77EE100 pKa = 3.94RR101 pKa = 11.84EE102 pKa = 4.25AYY104 pKa = 9.6GVHH107 pKa = 6.36FPSEE111 pKa = 4.36KK112 pKa = 9.28PTIDD116 pKa = 5.23LSDD119 pKa = 4.22LPATLQRR126 pKa = 11.84RR127 pKa = 11.84IFVDD131 pKa = 3.49ISNAFKK137 pKa = 11.27KK138 pKa = 10.28MMEE141 pKa = 4.44APSCQHH147 pKa = 5.7FPVMVRR153 pKa = 11.84GRR155 pKa = 11.84IEE157 pKa = 5.33LIPSSDD163 pKa = 3.39MAIHH167 pKa = 5.26QTPSSATTPEE177 pKa = 4.43SEE179 pKa = 5.14DD180 pKa = 3.99LPPSPVKK187 pKa = 10.12RR188 pKa = 11.84DD189 pKa = 3.33RR190 pKa = 11.84LLDD193 pKa = 3.35RR194 pKa = 11.84FKK196 pKa = 10.27IHH198 pKa = 6.7HH199 pKa = 7.21RR200 pKa = 11.84STQDD204 pKa = 3.31PLEE207 pKa = 4.23TSFVEE212 pKa = 4.13IFGSEE217 pKa = 3.94DD218 pKa = 2.96SARR221 pKa = 11.84AAWGDD226 pKa = 3.26GSLSYY231 pKa = 10.97KK232 pKa = 10.29EE233 pKa = 4.65LLILGLRR240 pKa = 11.84RR241 pKa = 11.84KK242 pKa = 10.1GVFNRR247 pKa = 11.84MRR249 pKa = 11.84IAYY252 pKa = 8.71DD253 pKa = 3.25LTPIFEE259 pKa = 4.36
Molecular weight: 29.24 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0D3R1T8|A0A0D3R1T8_9RHAB Phosphoprotein OS=Radi vesiculovirus OX=1972566 PE=3 SV=1
MM1 pKa = 7.6KK2 pKa = 10.18GFKK5 pKa = 10.85AMFSKK10 pKa = 10.63KK11 pKa = 9.62KK12 pKa = 9.46SKK14 pKa = 10.03KK15 pKa = 9.51GKK17 pKa = 9.14EE18 pKa = 3.62IKK20 pKa = 10.62AKK22 pKa = 10.84GKK24 pKa = 10.87GKK26 pKa = 10.24DD27 pKa = 3.16KK28 pKa = 11.02NKK30 pKa = 10.43QYY32 pKa = 11.24DD33 pKa = 3.59EE34 pKa = 4.7DD35 pKa = 4.2EE36 pKa = 4.53RR37 pKa = 11.84PPAYY41 pKa = 10.55FNVAPEE47 pKa = 4.24WGSASAPMFGYY58 pKa = 10.44DD59 pKa = 4.53EE60 pKa = 4.98YY61 pKa = 11.02IEE63 pKa = 4.43SQSSIQSLKK72 pKa = 9.45IQFRR76 pKa = 11.84YY77 pKa = 9.95SLEE80 pKa = 4.06VTVDD84 pKa = 3.3YY85 pKa = 10.75PFANFFEE92 pKa = 5.04ALQSMRR98 pKa = 11.84NWKK101 pKa = 9.38IDD103 pKa = 3.26YY104 pKa = 9.19TGYY107 pKa = 10.33QNKK110 pKa = 9.82KK111 pKa = 9.15SIYY114 pKa = 8.97NIIMMYY120 pKa = 8.01ATKK123 pKa = 10.43RR124 pKa = 11.84LRR126 pKa = 11.84AGPRR130 pKa = 11.84GLQGRR135 pKa = 11.84KK136 pKa = 5.82STEE139 pKa = 3.63YY140 pKa = 10.09SAEE143 pKa = 3.73GSGRR147 pKa = 11.84YY148 pKa = 6.75TLHH151 pKa = 7.24HH152 pKa = 7.3DD153 pKa = 3.92LGDD156 pKa = 3.64LSGMRR161 pKa = 11.84FAEE164 pKa = 4.15EE165 pKa = 3.84RR166 pKa = 11.84FVRR169 pKa = 11.84TWQDD173 pKa = 2.89QLRR176 pKa = 11.84RR177 pKa = 11.84GTINFYY183 pKa = 10.44VWLGEE188 pKa = 3.94TDD190 pKa = 4.28DD191 pKa = 4.77NSNLPPLICPGDD203 pKa = 3.6FKK205 pKa = 11.28DD206 pKa = 3.67ASEE209 pKa = 4.75FNMSCEE215 pKa = 3.64IMGIRR220 pKa = 11.84VEE222 pKa = 4.19VQPDD226 pKa = 3.56NQWVVKK232 pKa = 9.65EE233 pKa = 4.01AA234 pKa = 3.52
MM1 pKa = 7.6KK2 pKa = 10.18GFKK5 pKa = 10.85AMFSKK10 pKa = 10.63KK11 pKa = 9.62KK12 pKa = 9.46SKK14 pKa = 10.03KK15 pKa = 9.51GKK17 pKa = 9.14EE18 pKa = 3.62IKK20 pKa = 10.62AKK22 pKa = 10.84GKK24 pKa = 10.87GKK26 pKa = 10.24DD27 pKa = 3.16KK28 pKa = 11.02NKK30 pKa = 10.43QYY32 pKa = 11.24DD33 pKa = 3.59EE34 pKa = 4.7DD35 pKa = 4.2EE36 pKa = 4.53RR37 pKa = 11.84PPAYY41 pKa = 10.55FNVAPEE47 pKa = 4.24WGSASAPMFGYY58 pKa = 10.44DD59 pKa = 4.53EE60 pKa = 4.98YY61 pKa = 11.02IEE63 pKa = 4.43SQSSIQSLKK72 pKa = 9.45IQFRR76 pKa = 11.84YY77 pKa = 9.95SLEE80 pKa = 4.06VTVDD84 pKa = 3.3YY85 pKa = 10.75PFANFFEE92 pKa = 5.04ALQSMRR98 pKa = 11.84NWKK101 pKa = 9.38IDD103 pKa = 3.26YY104 pKa = 9.19TGYY107 pKa = 10.33QNKK110 pKa = 9.82KK111 pKa = 9.15SIYY114 pKa = 8.97NIIMMYY120 pKa = 8.01ATKK123 pKa = 10.43RR124 pKa = 11.84LRR126 pKa = 11.84AGPRR130 pKa = 11.84GLQGRR135 pKa = 11.84KK136 pKa = 5.82STEE139 pKa = 3.63YY140 pKa = 10.09SAEE143 pKa = 3.73GSGRR147 pKa = 11.84YY148 pKa = 6.75TLHH151 pKa = 7.24HH152 pKa = 7.3DD153 pKa = 3.92LGDD156 pKa = 3.64LSGMRR161 pKa = 11.84FAEE164 pKa = 4.15EE165 pKa = 3.84RR166 pKa = 11.84FVRR169 pKa = 11.84TWQDD173 pKa = 2.89QLRR176 pKa = 11.84RR177 pKa = 11.84GTINFYY183 pKa = 10.44VWLGEE188 pKa = 3.94TDD190 pKa = 4.28DD191 pKa = 4.77NSNLPPLICPGDD203 pKa = 3.6FKK205 pKa = 11.28DD206 pKa = 3.67ASEE209 pKa = 4.75FNMSCEE215 pKa = 3.64IMGIRR220 pKa = 11.84VEE222 pKa = 4.19VQPDD226 pKa = 3.56NQWVVKK232 pKa = 9.65EE233 pKa = 4.01AA234 pKa = 3.52
Molecular weight: 27.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3577 |
234 |
2100 |
715.4 |
81.16 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.284 ± 0.883 | 1.594 ± 0.241 |
6.039 ± 0.616 | 5.927 ± 0.58 |
4.389 ± 0.214 | 6.458 ± 0.443 |
2.572 ± 0.23 | 6.458 ± 0.651 |
5.563 ± 0.624 | 9.785 ± 1.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.544 ± 0.152 | 4.166 ± 0.381 |
4.501 ± 0.446 | 3.327 ± 0.342 |
6.346 ± 0.589 | 9.365 ± 0.483 |
5.088 ± 0.436 | 5.396 ± 0.593 |
2.041 ± 0.269 | 3.159 ± 0.279 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
