
Parahaliea aestuarii
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Cellvibrionales; Halieaceae; Parahaliea
Average proteome isoelectric point is 5.87
Get precalculated fractions of proteins
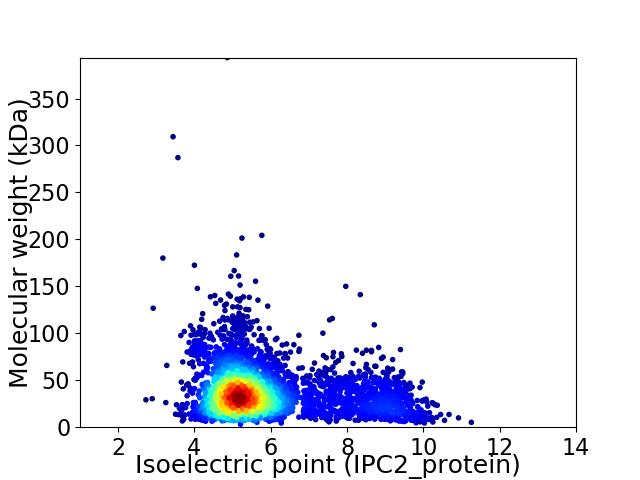
Virtual 2D-PAGE plot for 3815 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
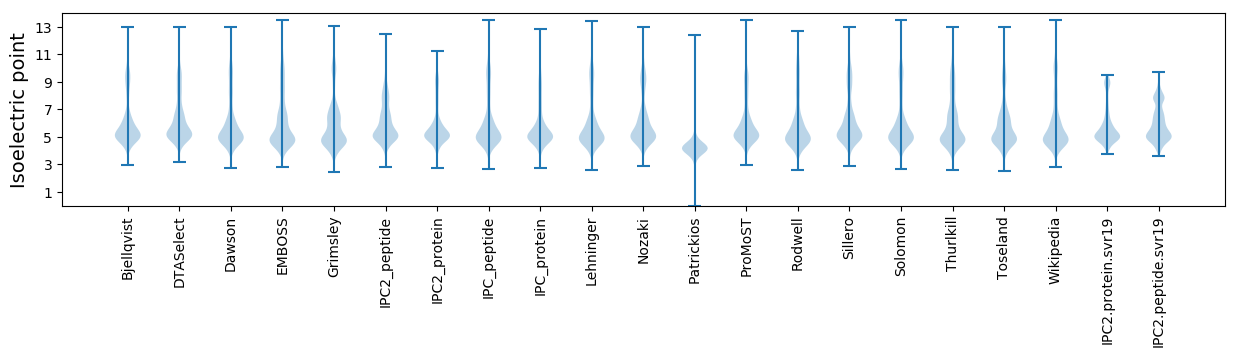
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5C9A0W4|A0A5C9A0W4_9GAMM LysR family transcriptional regulator OS=Parahaliea aestuarii OX=1852021 GN=FVW59_05175 PE=3 SV=1
MM1 pKa = 7.58NLQNYY6 pKa = 9.06NLDD9 pKa = 2.85IGYY12 pKa = 10.46ALGNGMVLRR21 pKa = 11.84SITGYY26 pKa = 11.01NDD28 pKa = 3.06TDD30 pKa = 4.2LEE32 pKa = 4.39ITSVPSAADD41 pKa = 3.15VFFRR45 pKa = 11.84QDD47 pKa = 2.5NRR49 pKa = 11.84MDD51 pKa = 3.9GDD53 pKa = 3.95FMQEE57 pKa = 3.87FRR59 pKa = 11.84LEE61 pKa = 4.17MNEE64 pKa = 4.15EE65 pKa = 3.97IAGLTGVASLFYY77 pKa = 11.03GDD79 pKa = 5.0FDD81 pKa = 3.65QHH83 pKa = 7.06SEE85 pKa = 4.22SYY87 pKa = 10.07ILYY90 pKa = 8.27YY91 pKa = 10.25QAFPVQDD98 pKa = 3.49GTFDD102 pKa = 3.53NSTEE106 pKa = 3.59TWAAYY111 pKa = 10.5ADD113 pKa = 3.55FRR115 pKa = 11.84YY116 pKa = 10.35RR117 pKa = 11.84LTDD120 pKa = 3.37SLSLLFGGRR129 pKa = 11.84YY130 pKa = 9.16QDD132 pKa = 3.22DD133 pKa = 4.06TVRR136 pKa = 11.84NQADD140 pKa = 3.63VEE142 pKa = 4.51SALSGGSQYY151 pKa = 11.38DD152 pKa = 3.81LEE154 pKa = 5.05SGFDD158 pKa = 3.38MFLPKK163 pKa = 10.48AGLSLDD169 pKa = 3.23ITDD172 pKa = 4.19NQTVAATASRR182 pKa = 11.84GYY184 pKa = 9.97RR185 pKa = 11.84QGFAEE190 pKa = 4.13RR191 pKa = 11.84RR192 pKa = 11.84LGSEE196 pKa = 3.87AGLVDD201 pKa = 3.98VDD203 pKa = 5.11PEE205 pKa = 4.35FVWAYY210 pKa = 9.09EE211 pKa = 3.45IAYY214 pKa = 9.93RR215 pKa = 11.84ISALNNRR222 pKa = 11.84LLLGANVFYY231 pKa = 11.11NDD233 pKa = 3.67YY234 pKa = 9.64TDD236 pKa = 3.55QQITVTNPDD245 pKa = 3.89FAPLTNTMNAGDD257 pKa = 4.14SEE259 pKa = 4.69SWGAEE264 pKa = 3.35IEE266 pKa = 4.21AQLAFDD272 pKa = 3.9NGLRR276 pKa = 11.84IYY278 pKa = 10.92GSLGLLEE285 pKa = 4.58TEE287 pKa = 4.99LGDD290 pKa = 6.04FPAEE294 pKa = 4.14DD295 pKa = 4.7CSNGSCDD302 pKa = 3.21GNQYY306 pKa = 10.85SEE308 pKa = 4.51APEE311 pKa = 4.08VTASLGGEE319 pKa = 4.01YY320 pKa = 10.18RR321 pKa = 11.84HH322 pKa = 7.04DD323 pKa = 3.6SGFFASLAASYY334 pKa = 10.2TDD336 pKa = 2.99SFYY339 pKa = 11.22RR340 pKa = 11.84SVDD343 pKa = 3.32NADD346 pKa = 3.82DD347 pKa = 4.75LEE349 pKa = 4.94VDD351 pKa = 3.45SSFVVDD357 pKa = 4.84AKK359 pKa = 10.43IGYY362 pKa = 9.4DD363 pKa = 3.36FGHH366 pKa = 6.25FRR368 pKa = 11.84VSAYY372 pKa = 10.57ANNVFDD378 pKa = 4.78EE379 pKa = 4.97EE380 pKa = 4.88YY381 pKa = 10.06LTGISNANSAFIGDD395 pKa = 3.9PRR397 pKa = 11.84AMGVEE402 pKa = 4.2VTAAFF407 pKa = 3.91
MM1 pKa = 7.58NLQNYY6 pKa = 9.06NLDD9 pKa = 2.85IGYY12 pKa = 10.46ALGNGMVLRR21 pKa = 11.84SITGYY26 pKa = 11.01NDD28 pKa = 3.06TDD30 pKa = 4.2LEE32 pKa = 4.39ITSVPSAADD41 pKa = 3.15VFFRR45 pKa = 11.84QDD47 pKa = 2.5NRR49 pKa = 11.84MDD51 pKa = 3.9GDD53 pKa = 3.95FMQEE57 pKa = 3.87FRR59 pKa = 11.84LEE61 pKa = 4.17MNEE64 pKa = 4.15EE65 pKa = 3.97IAGLTGVASLFYY77 pKa = 11.03GDD79 pKa = 5.0FDD81 pKa = 3.65QHH83 pKa = 7.06SEE85 pKa = 4.22SYY87 pKa = 10.07ILYY90 pKa = 8.27YY91 pKa = 10.25QAFPVQDD98 pKa = 3.49GTFDD102 pKa = 3.53NSTEE106 pKa = 3.59TWAAYY111 pKa = 10.5ADD113 pKa = 3.55FRR115 pKa = 11.84YY116 pKa = 10.35RR117 pKa = 11.84LTDD120 pKa = 3.37SLSLLFGGRR129 pKa = 11.84YY130 pKa = 9.16QDD132 pKa = 3.22DD133 pKa = 4.06TVRR136 pKa = 11.84NQADD140 pKa = 3.63VEE142 pKa = 4.51SALSGGSQYY151 pKa = 11.38DD152 pKa = 3.81LEE154 pKa = 5.05SGFDD158 pKa = 3.38MFLPKK163 pKa = 10.48AGLSLDD169 pKa = 3.23ITDD172 pKa = 4.19NQTVAATASRR182 pKa = 11.84GYY184 pKa = 9.97RR185 pKa = 11.84QGFAEE190 pKa = 4.13RR191 pKa = 11.84RR192 pKa = 11.84LGSEE196 pKa = 3.87AGLVDD201 pKa = 3.98VDD203 pKa = 5.11PEE205 pKa = 4.35FVWAYY210 pKa = 9.09EE211 pKa = 3.45IAYY214 pKa = 9.93RR215 pKa = 11.84ISALNNRR222 pKa = 11.84LLLGANVFYY231 pKa = 11.11NDD233 pKa = 3.67YY234 pKa = 9.64TDD236 pKa = 3.55QQITVTNPDD245 pKa = 3.89FAPLTNTMNAGDD257 pKa = 4.14SEE259 pKa = 4.69SWGAEE264 pKa = 3.35IEE266 pKa = 4.21AQLAFDD272 pKa = 3.9NGLRR276 pKa = 11.84IYY278 pKa = 10.92GSLGLLEE285 pKa = 4.58TEE287 pKa = 4.99LGDD290 pKa = 6.04FPAEE294 pKa = 4.14DD295 pKa = 4.7CSNGSCDD302 pKa = 3.21GNQYY306 pKa = 10.85SEE308 pKa = 4.51APEE311 pKa = 4.08VTASLGGEE319 pKa = 4.01YY320 pKa = 10.18RR321 pKa = 11.84HH322 pKa = 7.04DD323 pKa = 3.6SGFFASLAASYY334 pKa = 10.2TDD336 pKa = 2.99SFYY339 pKa = 11.22RR340 pKa = 11.84SVDD343 pKa = 3.32NADD346 pKa = 3.82DD347 pKa = 4.75LEE349 pKa = 4.94VDD351 pKa = 3.45SSFVVDD357 pKa = 4.84AKK359 pKa = 10.43IGYY362 pKa = 9.4DD363 pKa = 3.36FGHH366 pKa = 6.25FRR368 pKa = 11.84VSAYY372 pKa = 10.57ANNVFDD378 pKa = 4.78EE379 pKa = 4.97EE380 pKa = 4.88YY381 pKa = 10.06LTGISNANSAFIGDD395 pKa = 3.9PRR397 pKa = 11.84AMGVEE402 pKa = 4.2VTAAFF407 pKa = 3.91
Molecular weight: 44.85 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5C8ZY70|A0A5C8ZY70_9GAMM Type I secretion system permease/ATPase OS=Parahaliea aestuarii OX=1852021 GN=FVW59_06940 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVIKK11 pKa = 10.52RR12 pKa = 11.84KK13 pKa = 8.26RR14 pKa = 11.84THH16 pKa = 5.92GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.24NGRR28 pKa = 11.84LVLSRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.02GRR39 pKa = 11.84KK40 pKa = 8.77RR41 pKa = 11.84LAAA44 pKa = 4.42
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVIKK11 pKa = 10.52RR12 pKa = 11.84KK13 pKa = 8.26RR14 pKa = 11.84THH16 pKa = 5.92GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.24NGRR28 pKa = 11.84LVLSRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.02GRR39 pKa = 11.84KK40 pKa = 8.77RR41 pKa = 11.84LAAA44 pKa = 4.42
Molecular weight: 5.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1363449 |
28 |
3668 |
357.4 |
39.1 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.236 ± 0.043 | 1.054 ± 0.014 |
5.941 ± 0.03 | 6.245 ± 0.035 |
3.609 ± 0.025 | 8.371 ± 0.043 |
2.134 ± 0.017 | 4.577 ± 0.029 |
2.688 ± 0.032 | 10.908 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.264 ± 0.021 | 3.007 ± 0.026 |
4.873 ± 0.027 | 4.07 ± 0.03 |
6.815 ± 0.039 | 5.986 ± 0.031 |
4.869 ± 0.029 | 7.146 ± 0.034 |
1.41 ± 0.016 | 2.797 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
