
Icoaraci virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Ellioviricetes; Bunyavirales; Phenuiviridae; Phlebovirus; Icoaraci phlebovirus
Average proteome isoelectric point is 6.47
Get precalculated fractions of proteins
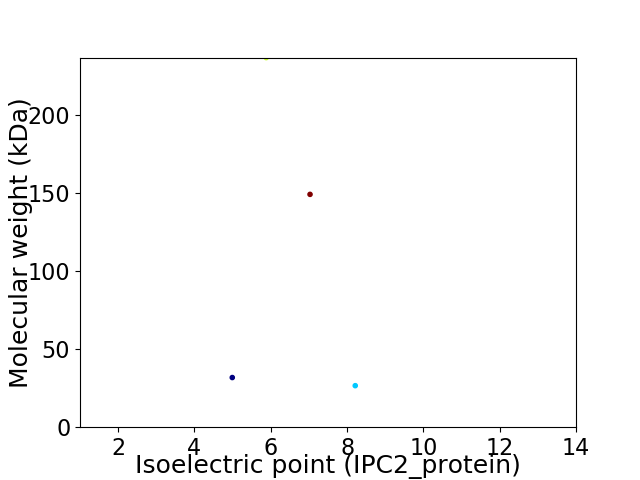
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8D7V5|A0A4P8D7V5_9VIRU Glycoprotein OS=Icoaraci virus OX=426790 PE=4 SV=1
MM1 pKa = 7.59NIMNFIYY8 pKa = 10.03EE9 pKa = 4.07MPIILSYY16 pKa = 11.41GGMGNPEE23 pKa = 4.38AEE25 pKa = 4.2VGYY28 pKa = 10.43YY29 pKa = 9.77PYY31 pKa = 10.96EE32 pKa = 4.1GFTTYY37 pKa = 10.03RR38 pKa = 11.84VSVHH42 pKa = 6.83HH43 pKa = 7.21DD44 pKa = 3.12MEE46 pKa = 6.03IPVGTYY52 pKa = 9.44RR53 pKa = 11.84QVMDD57 pKa = 3.89HH58 pKa = 7.22RR59 pKa = 11.84STLEE63 pKa = 3.67DD64 pKa = 4.51FYY66 pKa = 11.75SRR68 pKa = 11.84GHH70 pKa = 6.38FPFRR74 pKa = 11.84WGPGSFNSRR83 pKa = 11.84VEE85 pKa = 4.12NPTCSDD91 pKa = 3.24FDD93 pKa = 3.76EE94 pKa = 4.45MLRR97 pKa = 11.84EE98 pKa = 4.45LMLFPNEE105 pKa = 3.7AYY107 pKa = 8.84TRR109 pKa = 11.84SFLPNISEE117 pKa = 4.66AISWPLGYY125 pKa = 9.35PCSKK129 pKa = 10.49FIQLSNEE136 pKa = 4.07QKK138 pKa = 10.33SQLTLRR144 pKa = 11.84SWKK147 pKa = 10.1SAAATCILRR156 pKa = 11.84MNPWVSTFDD165 pKa = 3.38QAFVEE170 pKa = 4.12AHH172 pKa = 6.3KK173 pKa = 10.44MILKK177 pKa = 9.46EE178 pKa = 3.82ATNRR182 pKa = 11.84GLSRR186 pKa = 11.84MLFPGYY192 pKa = 10.72DD193 pKa = 3.77LIKK196 pKa = 10.41EE197 pKa = 4.17VAIVQLVRR205 pKa = 11.84AMNALDD211 pKa = 3.61IDD213 pKa = 4.04MTYY216 pKa = 11.12SRR218 pKa = 11.84IPTVLTNMLEE228 pKa = 3.76KK229 pKa = 10.49HH230 pKa = 6.19KK231 pKa = 11.17NVFEE235 pKa = 4.21NEE237 pKa = 3.91NPDD240 pKa = 3.86LLGNRR245 pKa = 11.84KK246 pKa = 8.96WIPSDD251 pKa = 3.52DD252 pKa = 3.23TWFEE256 pKa = 3.72VLNYY260 pKa = 10.0HH261 pKa = 6.78GEE263 pKa = 4.42SEE265 pKa = 4.81FDD267 pKa = 3.26TSSDD271 pKa = 2.82SDD273 pKa = 3.7AAA275 pKa = 3.95
MM1 pKa = 7.59NIMNFIYY8 pKa = 10.03EE9 pKa = 4.07MPIILSYY16 pKa = 11.41GGMGNPEE23 pKa = 4.38AEE25 pKa = 4.2VGYY28 pKa = 10.43YY29 pKa = 9.77PYY31 pKa = 10.96EE32 pKa = 4.1GFTTYY37 pKa = 10.03RR38 pKa = 11.84VSVHH42 pKa = 6.83HH43 pKa = 7.21DD44 pKa = 3.12MEE46 pKa = 6.03IPVGTYY52 pKa = 9.44RR53 pKa = 11.84QVMDD57 pKa = 3.89HH58 pKa = 7.22RR59 pKa = 11.84STLEE63 pKa = 3.67DD64 pKa = 4.51FYY66 pKa = 11.75SRR68 pKa = 11.84GHH70 pKa = 6.38FPFRR74 pKa = 11.84WGPGSFNSRR83 pKa = 11.84VEE85 pKa = 4.12NPTCSDD91 pKa = 3.24FDD93 pKa = 3.76EE94 pKa = 4.45MLRR97 pKa = 11.84EE98 pKa = 4.45LMLFPNEE105 pKa = 3.7AYY107 pKa = 8.84TRR109 pKa = 11.84SFLPNISEE117 pKa = 4.66AISWPLGYY125 pKa = 9.35PCSKK129 pKa = 10.49FIQLSNEE136 pKa = 4.07QKK138 pKa = 10.33SQLTLRR144 pKa = 11.84SWKK147 pKa = 10.1SAAATCILRR156 pKa = 11.84MNPWVSTFDD165 pKa = 3.38QAFVEE170 pKa = 4.12AHH172 pKa = 6.3KK173 pKa = 10.44MILKK177 pKa = 9.46EE178 pKa = 3.82ATNRR182 pKa = 11.84GLSRR186 pKa = 11.84MLFPGYY192 pKa = 10.72DD193 pKa = 3.77LIKK196 pKa = 10.41EE197 pKa = 4.17VAIVQLVRR205 pKa = 11.84AMNALDD211 pKa = 3.61IDD213 pKa = 4.04MTYY216 pKa = 11.12SRR218 pKa = 11.84IPTVLTNMLEE228 pKa = 3.76KK229 pKa = 10.49HH230 pKa = 6.19KK231 pKa = 11.17NVFEE235 pKa = 4.21NEE237 pKa = 3.91NPDD240 pKa = 3.86LLGNRR245 pKa = 11.84KK246 pKa = 8.96WIPSDD251 pKa = 3.52DD252 pKa = 3.23TWFEE256 pKa = 3.72VLNYY260 pKa = 10.0HH261 pKa = 6.78GEE263 pKa = 4.42SEE265 pKa = 4.81FDD267 pKa = 3.26TSSDD271 pKa = 2.82SDD273 pKa = 3.7AAA275 pKa = 3.95
Molecular weight: 31.8 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8D7V8|A0A4P8D7V8_9VIRU Nucleoprotein OS=Icoaraci virus OX=426790 PE=3 SV=1
MM1 pKa = 7.54TDD3 pKa = 3.26YY4 pKa = 11.58ARR6 pKa = 11.84IAVEE10 pKa = 4.26FSGEE14 pKa = 3.98AVNLAEE20 pKa = 4.11IQGWVADD27 pKa = 4.23FAYY30 pKa = 10.43QGFDD34 pKa = 2.65ARR36 pKa = 11.84RR37 pKa = 11.84IVEE40 pKa = 4.28LVQQKK45 pKa = 10.65GGATWKK51 pKa = 10.63DD52 pKa = 3.68DD53 pKa = 3.7VKK55 pKa = 11.23MMIVLCLTRR64 pKa = 11.84GNKK67 pKa = 5.97PTKK70 pKa = 9.31MVEE73 pKa = 3.9KK74 pKa = 9.51MSPEE78 pKa = 3.86GKK80 pKa = 10.01VKK82 pKa = 10.44VNRR85 pKa = 11.84LISTYY90 pKa = 9.01GLKK93 pKa = 10.39SGNPGRR99 pKa = 11.84DD100 pKa = 3.8DD101 pKa = 3.22ITLSRR106 pKa = 11.84VAAAFAGWTCQALNVLHH123 pKa = 6.48PHH125 pKa = 6.59LPVSGTTMDD134 pKa = 5.36AISPNYY140 pKa = 8.24PRR142 pKa = 11.84AMMHH146 pKa = 6.41PCFAGLVDD154 pKa = 3.47PTIPAEE160 pKa = 4.04YY161 pKa = 10.1CQMIVDD167 pKa = 4.26AMSVFLIQFSRR178 pKa = 11.84TINKK182 pKa = 9.32SLRR185 pKa = 11.84GQPKK189 pKa = 7.92EE190 pKa = 3.77AVVEE194 pKa = 4.45SFVQPMQAAMASSFITPAEE213 pKa = 3.96RR214 pKa = 11.84RR215 pKa = 11.84KK216 pKa = 10.83LMIALGIVDD225 pKa = 5.03ANGKK229 pKa = 9.38PSANVAAAAAAYY241 pKa = 7.94PKK243 pKa = 10.63VII245 pKa = 4.17
MM1 pKa = 7.54TDD3 pKa = 3.26YY4 pKa = 11.58ARR6 pKa = 11.84IAVEE10 pKa = 4.26FSGEE14 pKa = 3.98AVNLAEE20 pKa = 4.11IQGWVADD27 pKa = 4.23FAYY30 pKa = 10.43QGFDD34 pKa = 2.65ARR36 pKa = 11.84RR37 pKa = 11.84IVEE40 pKa = 4.28LVQQKK45 pKa = 10.65GGATWKK51 pKa = 10.63DD52 pKa = 3.68DD53 pKa = 3.7VKK55 pKa = 11.23MMIVLCLTRR64 pKa = 11.84GNKK67 pKa = 5.97PTKK70 pKa = 9.31MVEE73 pKa = 3.9KK74 pKa = 9.51MSPEE78 pKa = 3.86GKK80 pKa = 10.01VKK82 pKa = 10.44VNRR85 pKa = 11.84LISTYY90 pKa = 9.01GLKK93 pKa = 10.39SGNPGRR99 pKa = 11.84DD100 pKa = 3.8DD101 pKa = 3.22ITLSRR106 pKa = 11.84VAAAFAGWTCQALNVLHH123 pKa = 6.48PHH125 pKa = 6.59LPVSGTTMDD134 pKa = 5.36AISPNYY140 pKa = 8.24PRR142 pKa = 11.84AMMHH146 pKa = 6.41PCFAGLVDD154 pKa = 3.47PTIPAEE160 pKa = 4.04YY161 pKa = 10.1CQMIVDD167 pKa = 4.26AMSVFLIQFSRR178 pKa = 11.84TINKK182 pKa = 9.32SLRR185 pKa = 11.84GQPKK189 pKa = 7.92EE190 pKa = 3.77AVVEE194 pKa = 4.45SFVQPMQAAMASSFITPAEE213 pKa = 3.96RR214 pKa = 11.84RR215 pKa = 11.84KK216 pKa = 10.83LMIALGIVDD225 pKa = 5.03ANGKK229 pKa = 9.38PSANVAAAAAAYY241 pKa = 7.94PKK243 pKa = 10.63VII245 pKa = 4.17
Molecular weight: 26.6 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3958 |
245 |
2084 |
989.5 |
111.06 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.584 ± 0.996 | 2.501 ± 0.79 |
5.508 ± 0.378 | 6.998 ± 0.627 |
4.219 ± 0.672 | 6.443 ± 0.644 |
2.35 ± 0.197 | 6.064 ± 0.117 |
6.771 ± 0.754 | 8.59 ± 0.486 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.487 ± 0.473 | 4.169 ± 0.413 |
4.548 ± 0.404 | 3.032 ± 0.156 |
5.154 ± 0.111 | 8.818 ± 0.618 |
5.104 ± 0.065 | 6.695 ± 0.425 |
1.289 ± 0.168 | 2.678 ± 0.357 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
