
Cochliobolus sativus (strain ND90Pr / ATCC 201652) (Common root rot and spot blotch fungus) (Bipolaris sorokiniana)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; dothideomyceta; Dothideomycetes; Pleosporomycetidae; Pleosporales; Pleosporineae; Pleosporaceae; Bipolaris; Bipolaris sorokiniana
Average proteome isoelectric point is 6.74
Get precalculated fractions of proteins
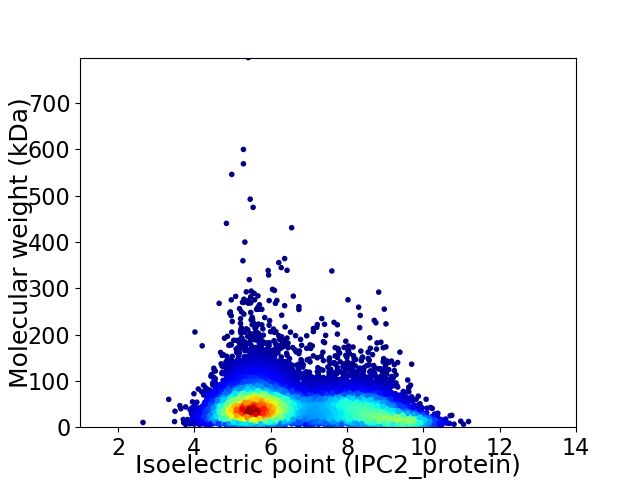
Virtual 2D-PAGE plot for 12174 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions



Protein with the lowest isoelectric point:
>tr|M2SVJ7|M2SVJ7_COCSN Fatty acid hydroxylase domain-containing protein OS=Cochliobolus sativus (strain ND90Pr / ATCC 201652) OX=665912 GN=COCSADRAFT_163448 PE=4 SV=1
MM1 pKa = 8.05DD2 pKa = 4.72LHH4 pKa = 8.59SDD6 pKa = 4.46DD7 pKa = 6.4GILGYY12 pKa = 10.81NDD14 pKa = 3.75GDD16 pKa = 3.6IEE18 pKa = 5.83LDD20 pKa = 3.7LEE22 pKa = 4.6PPPSYY27 pKa = 11.26GPDD30 pKa = 3.49DD31 pKa = 4.39DD32 pKa = 5.92VSIGDD37 pKa = 3.66AASANGMDD45 pKa = 4.12AQTAPGGDD53 pKa = 3.5NDD55 pKa = 5.76DD56 pKa = 4.09YY57 pKa = 11.43MVDD60 pKa = 3.38QEE62 pKa = 5.34DD63 pKa = 4.47VIEE66 pKa = 4.47EE67 pKa = 4.35DD68 pKa = 4.75YY69 pKa = 11.23SYY71 pKa = 11.43QDD73 pKa = 2.8QDD75 pKa = 2.83VDD77 pKa = 3.47VDD79 pKa = 3.62VDD81 pKa = 3.59VDD83 pKa = 3.74VDD85 pKa = 3.52VDD87 pKa = 4.04ADD89 pKa = 3.98AEE91 pKa = 4.37QPAATDD97 pKa = 4.16TIATQAEE104 pKa = 4.54AEE106 pKa = 4.38VQDD109 pKa = 4.77EE110 pKa = 4.35DD111 pKa = 4.27LLDD114 pKa = 3.92YY115 pKa = 11.35SEE117 pKa = 5.71EE118 pKa = 4.48DD119 pKa = 4.21DD120 pKa = 5.79DD121 pKa = 4.72YY122 pKa = 11.79HH123 pKa = 6.71DD124 pKa = 3.83TALRR128 pKa = 11.84SPWLQQHH135 pKa = 7.22DD136 pKa = 4.29SMQDD140 pKa = 3.04EE141 pKa = 5.07PGPQNEE147 pKa = 4.2PSQAQDD153 pKa = 3.21APVQQDD159 pKa = 4.46EE160 pKa = 4.91IPADD164 pKa = 4.0LQALMSTNTSKK175 pKa = 10.97NPQSPHH181 pKa = 5.08AHH183 pKa = 6.82PDD185 pKa = 3.33VVADD189 pKa = 5.09GSTKK193 pKa = 10.65GNNNEE198 pKa = 3.66NSPRR202 pKa = 11.84ARR204 pKa = 11.84SADD207 pKa = 3.62PNEE210 pKa = 4.47EE211 pKa = 3.95DD212 pKa = 4.06AQTNGDD218 pKa = 4.14DD219 pKa = 4.88GGITLKK225 pKa = 10.84DD226 pKa = 3.62QEE228 pKa = 4.45EE229 pKa = 4.32HH230 pKa = 6.7SSAEE234 pKa = 3.98SGHH237 pKa = 5.49NHH239 pKa = 5.76MEE241 pKa = 4.49PPSHH245 pKa = 6.08GHH247 pKa = 7.0DD248 pKa = 3.16EE249 pKa = 4.23SAQVDD254 pKa = 3.8EE255 pKa = 5.22HH256 pKa = 7.83EE257 pKa = 5.11EE258 pKa = 4.05DD259 pKa = 3.44TSFQQLPPVTVNCMGEE275 pKa = 4.2EE276 pKa = 3.65LWLFKK281 pKa = 10.76QHH283 pKa = 7.33DD284 pKa = 4.37YY285 pKa = 11.41EE286 pKa = 5.65NSGDD290 pKa = 3.59WLLADD295 pKa = 3.56VSVAKK300 pKa = 10.05TSMSDD305 pKa = 3.26LFQTCRR311 pKa = 11.84SALGHH316 pKa = 5.83EE317 pKa = 4.78VLNGMEE323 pKa = 3.48IGFRR327 pKa = 11.84FDD329 pKa = 3.73HH330 pKa = 5.78FQNMEE335 pKa = 4.12LFEE338 pKa = 5.04EE339 pKa = 4.35NTACVAISLEE349 pKa = 4.03RR350 pKa = 11.84LVGYY354 pKa = 7.91YY355 pKa = 9.9HH356 pKa = 7.65ALHH359 pKa = 6.29EE360 pKa = 4.22QDD362 pKa = 3.63
MM1 pKa = 8.05DD2 pKa = 4.72LHH4 pKa = 8.59SDD6 pKa = 4.46DD7 pKa = 6.4GILGYY12 pKa = 10.81NDD14 pKa = 3.75GDD16 pKa = 3.6IEE18 pKa = 5.83LDD20 pKa = 3.7LEE22 pKa = 4.6PPPSYY27 pKa = 11.26GPDD30 pKa = 3.49DD31 pKa = 4.39DD32 pKa = 5.92VSIGDD37 pKa = 3.66AASANGMDD45 pKa = 4.12AQTAPGGDD53 pKa = 3.5NDD55 pKa = 5.76DD56 pKa = 4.09YY57 pKa = 11.43MVDD60 pKa = 3.38QEE62 pKa = 5.34DD63 pKa = 4.47VIEE66 pKa = 4.47EE67 pKa = 4.35DD68 pKa = 4.75YY69 pKa = 11.23SYY71 pKa = 11.43QDD73 pKa = 2.8QDD75 pKa = 2.83VDD77 pKa = 3.47VDD79 pKa = 3.62VDD81 pKa = 3.59VDD83 pKa = 3.74VDD85 pKa = 3.52VDD87 pKa = 4.04ADD89 pKa = 3.98AEE91 pKa = 4.37QPAATDD97 pKa = 4.16TIATQAEE104 pKa = 4.54AEE106 pKa = 4.38VQDD109 pKa = 4.77EE110 pKa = 4.35DD111 pKa = 4.27LLDD114 pKa = 3.92YY115 pKa = 11.35SEE117 pKa = 5.71EE118 pKa = 4.48DD119 pKa = 4.21DD120 pKa = 5.79DD121 pKa = 4.72YY122 pKa = 11.79HH123 pKa = 6.71DD124 pKa = 3.83TALRR128 pKa = 11.84SPWLQQHH135 pKa = 7.22DD136 pKa = 4.29SMQDD140 pKa = 3.04EE141 pKa = 5.07PGPQNEE147 pKa = 4.2PSQAQDD153 pKa = 3.21APVQQDD159 pKa = 4.46EE160 pKa = 4.91IPADD164 pKa = 4.0LQALMSTNTSKK175 pKa = 10.97NPQSPHH181 pKa = 5.08AHH183 pKa = 6.82PDD185 pKa = 3.33VVADD189 pKa = 5.09GSTKK193 pKa = 10.65GNNNEE198 pKa = 3.66NSPRR202 pKa = 11.84ARR204 pKa = 11.84SADD207 pKa = 3.62PNEE210 pKa = 4.47EE211 pKa = 3.95DD212 pKa = 4.06AQTNGDD218 pKa = 4.14DD219 pKa = 4.88GGITLKK225 pKa = 10.84DD226 pKa = 3.62QEE228 pKa = 4.45EE229 pKa = 4.32HH230 pKa = 6.7SSAEE234 pKa = 3.98SGHH237 pKa = 5.49NHH239 pKa = 5.76MEE241 pKa = 4.49PPSHH245 pKa = 6.08GHH247 pKa = 7.0DD248 pKa = 3.16EE249 pKa = 4.23SAQVDD254 pKa = 3.8EE255 pKa = 5.22HH256 pKa = 7.83EE257 pKa = 5.11EE258 pKa = 4.05DD259 pKa = 3.44TSFQQLPPVTVNCMGEE275 pKa = 4.2EE276 pKa = 3.65LWLFKK281 pKa = 10.76QHH283 pKa = 7.33DD284 pKa = 4.37YY285 pKa = 11.41EE286 pKa = 5.65NSGDD290 pKa = 3.59WLLADD295 pKa = 3.56VSVAKK300 pKa = 10.05TSMSDD305 pKa = 3.26LFQTCRR311 pKa = 11.84SALGHH316 pKa = 5.83EE317 pKa = 4.78VLNGMEE323 pKa = 3.48IGFRR327 pKa = 11.84FDD329 pKa = 3.73HH330 pKa = 5.78FQNMEE335 pKa = 4.12LFEE338 pKa = 5.04EE339 pKa = 4.35NTACVAISLEE349 pKa = 4.03RR350 pKa = 11.84LVGYY354 pKa = 7.91YY355 pKa = 9.9HH356 pKa = 7.65ALHH359 pKa = 6.29EE360 pKa = 4.22QDD362 pKa = 3.63
Molecular weight: 39.86 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|M2TEI1|M2TEI1_COCSN TauD domain-containing protein (Fragment) OS=Cochliobolus sativus (strain ND90Pr / ATCC 201652) OX=665912 GN=COCSADRAFT_81895 PE=4 SV=1
MM1 pKa = 7.31LHH3 pKa = 6.5HH4 pKa = 7.21APHH7 pKa = 6.42TMHH10 pKa = 6.84ATLLSSLRR18 pKa = 11.84PSPPSSSPFLPRR30 pKa = 11.84AQRR33 pKa = 11.84IHH35 pKa = 6.15SLRR38 pKa = 11.84ARR40 pKa = 11.84TTSPRR45 pKa = 11.84THH47 pKa = 6.92LPLSFPSSAPLHH59 pKa = 4.87STIPKK64 pKa = 10.14LLLLHH69 pKa = 6.3PTLTIPLAPSTNPLWPRR86 pKa = 11.84LPLRR90 pKa = 11.84TPFPFPNPLSNVRR103 pKa = 11.84TMHH106 pKa = 7.33RR107 pKa = 11.84ITVV110 pKa = 3.42
MM1 pKa = 7.31LHH3 pKa = 6.5HH4 pKa = 7.21APHH7 pKa = 6.42TMHH10 pKa = 6.84ATLLSSLRR18 pKa = 11.84PSPPSSSPFLPRR30 pKa = 11.84AQRR33 pKa = 11.84IHH35 pKa = 6.15SLRR38 pKa = 11.84ARR40 pKa = 11.84TTSPRR45 pKa = 11.84THH47 pKa = 6.92LPLSFPSSAPLHH59 pKa = 4.87STIPKK64 pKa = 10.14LLLLHH69 pKa = 6.3PTLTIPLAPSTNPLWPRR86 pKa = 11.84LPLRR90 pKa = 11.84TPFPFPNPLSNVRR103 pKa = 11.84TMHH106 pKa = 7.33RR107 pKa = 11.84ITVV110 pKa = 3.42
Molecular weight: 12.28 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5418225 |
49 |
7203 |
445.1 |
49.32 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.695 ± 0.022 | 1.293 ± 0.009 |
5.569 ± 0.017 | 6.099 ± 0.022 |
3.678 ± 0.013 | 6.681 ± 0.022 |
2.474 ± 0.01 | 4.937 ± 0.015 |
5.124 ± 0.019 | 8.674 ± 0.023 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.243 ± 0.009 | 3.788 ± 0.013 |
6.096 ± 0.024 | 4.176 ± 0.018 |
5.973 ± 0.02 | 8.094 ± 0.027 |
6.072 ± 0.016 | 6.018 ± 0.016 |
1.467 ± 0.008 | 2.848 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
