
Frigoribacterium sp. RIT-PI-h
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Frigoribacterium; unclassified Frigoribacterium
Average proteome isoelectric point is 6.07
Get precalculated fractions of proteins
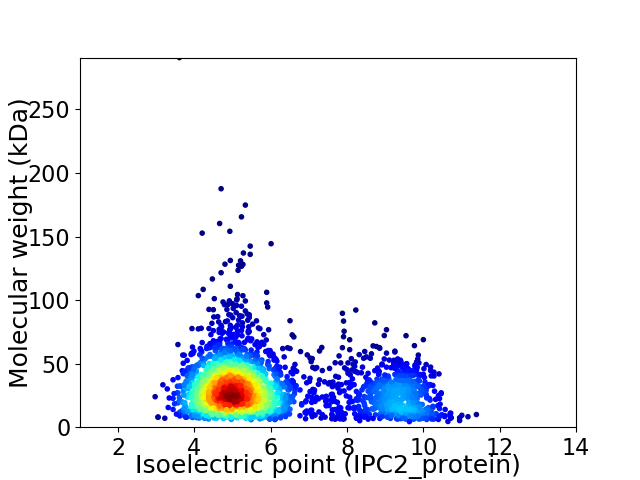
Virtual 2D-PAGE plot for 2815 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0N1M1S2|A0A0N1M1S2_9MICO Histidyl-tRNA synthetase OS=Frigoribacterium sp. RIT-PI-h OX=1690245 GN=AEQ27_13795 PE=3 SV=1
MM1 pKa = 7.25IRR3 pKa = 11.84SSTALRR9 pKa = 11.84ALALAGAASVLLAACSTASPSDD31 pKa = 3.87DD32 pKa = 3.49ASSSSSAKK40 pKa = 10.03ADD42 pKa = 3.57PAAKK46 pKa = 10.19CEE48 pKa = 4.09VGTPSTDD55 pKa = 2.92PFAIGTMLPLTGTLAYY71 pKa = 10.17LGPPAIAGANLAISDD86 pKa = 3.69INAAGGVLDD95 pKa = 4.16QPAEE99 pKa = 4.07ISTSTDD105 pKa = 3.02SGDD108 pKa = 3.94SNDD111 pKa = 3.62MTVSSNAATEE121 pKa = 4.94LIDD124 pKa = 3.98AKK126 pKa = 11.2VPVVLGAEE134 pKa = 4.37SSSVTLNVVDD144 pKa = 5.0QLTSNCIIEE153 pKa = 4.26ISPANTASSLSGYY166 pKa = 9.56SSYY169 pKa = 11.47YY170 pKa = 10.15YY171 pKa = 9.43RR172 pKa = 11.84TAPPDD177 pKa = 3.69SVQGSALGQQITADD191 pKa = 3.53GNANVAFLVFNDD203 pKa = 4.01TYY205 pKa = 11.06GTGLRR210 pKa = 11.84DD211 pKa = 3.51STQKK215 pKa = 10.78SIEE218 pKa = 4.25SSGGTVVYY226 pKa = 10.57GGTGKK231 pKa = 10.98GEE233 pKa = 4.21EE234 pKa = 4.4FPPGQTTFSSEE245 pKa = 3.56VSAAIAAEE253 pKa = 3.84PDD255 pKa = 4.02AIVILAFDD263 pKa = 3.92EE264 pKa = 4.59TKK266 pKa = 10.85SIVPEE271 pKa = 4.35LKK273 pKa = 10.26SQGWDD278 pKa = 3.06MSKK281 pKa = 11.1VYY283 pKa = 10.34MSDD286 pKa = 3.16GNTADD291 pKa = 3.5YY292 pKa = 11.13SADD295 pKa = 3.69FEE297 pKa = 5.07PGTLEE302 pKa = 4.11GAKK305 pKa = 9.21GTIPGADD312 pKa = 3.07ASADD316 pKa = 3.76FKK318 pKa = 11.5SKK320 pKa = 10.55LVAGYY325 pKa = 8.86QASEE329 pKa = 4.14GGEE332 pKa = 3.91LADD335 pKa = 4.23FSYY338 pKa = 11.03AAEE341 pKa = 4.53SYY343 pKa = 11.03DD344 pKa = 4.52AVILTALAAQAGGGTDD360 pKa = 3.37AGTIQANMAAVSGANGGTEE379 pKa = 3.95CTTYY383 pKa = 11.36ADD385 pKa = 4.44CKK387 pKa = 9.2TALDD391 pKa = 4.24AGDD394 pKa = 5.21DD395 pKa = 3.38IHH397 pKa = 6.17YY398 pKa = 7.46TGPSGIGPFNDD409 pKa = 3.4NNDD412 pKa = 3.67PSSASIGVYY421 pKa = 9.69TFDD424 pKa = 4.74ADD426 pKa = 3.77NKK428 pKa = 10.01PVYY431 pKa = 8.96QTGIEE436 pKa = 4.33GKK438 pKa = 10.17VEE440 pKa = 3.79
MM1 pKa = 7.25IRR3 pKa = 11.84SSTALRR9 pKa = 11.84ALALAGAASVLLAACSTASPSDD31 pKa = 3.87DD32 pKa = 3.49ASSSSSAKK40 pKa = 10.03ADD42 pKa = 3.57PAAKK46 pKa = 10.19CEE48 pKa = 4.09VGTPSTDD55 pKa = 2.92PFAIGTMLPLTGTLAYY71 pKa = 10.17LGPPAIAGANLAISDD86 pKa = 3.69INAAGGVLDD95 pKa = 4.16QPAEE99 pKa = 4.07ISTSTDD105 pKa = 3.02SGDD108 pKa = 3.94SNDD111 pKa = 3.62MTVSSNAATEE121 pKa = 4.94LIDD124 pKa = 3.98AKK126 pKa = 11.2VPVVLGAEE134 pKa = 4.37SSSVTLNVVDD144 pKa = 5.0QLTSNCIIEE153 pKa = 4.26ISPANTASSLSGYY166 pKa = 9.56SSYY169 pKa = 11.47YY170 pKa = 10.15YY171 pKa = 9.43RR172 pKa = 11.84TAPPDD177 pKa = 3.69SVQGSALGQQITADD191 pKa = 3.53GNANVAFLVFNDD203 pKa = 4.01TYY205 pKa = 11.06GTGLRR210 pKa = 11.84DD211 pKa = 3.51STQKK215 pKa = 10.78SIEE218 pKa = 4.25SSGGTVVYY226 pKa = 10.57GGTGKK231 pKa = 10.98GEE233 pKa = 4.21EE234 pKa = 4.4FPPGQTTFSSEE245 pKa = 3.56VSAAIAAEE253 pKa = 3.84PDD255 pKa = 4.02AIVILAFDD263 pKa = 3.92EE264 pKa = 4.59TKK266 pKa = 10.85SIVPEE271 pKa = 4.35LKK273 pKa = 10.26SQGWDD278 pKa = 3.06MSKK281 pKa = 11.1VYY283 pKa = 10.34MSDD286 pKa = 3.16GNTADD291 pKa = 3.5YY292 pKa = 11.13SADD295 pKa = 3.69FEE297 pKa = 5.07PGTLEE302 pKa = 4.11GAKK305 pKa = 9.21GTIPGADD312 pKa = 3.07ASADD316 pKa = 3.76FKK318 pKa = 11.5SKK320 pKa = 10.55LVAGYY325 pKa = 8.86QASEE329 pKa = 4.14GGEE332 pKa = 3.91LADD335 pKa = 4.23FSYY338 pKa = 11.03AAEE341 pKa = 4.53SYY343 pKa = 11.03DD344 pKa = 4.52AVILTALAAQAGGGTDD360 pKa = 3.37AGTIQANMAAVSGANGGTEE379 pKa = 3.95CTTYY383 pKa = 11.36ADD385 pKa = 4.44CKK387 pKa = 9.2TALDD391 pKa = 4.24AGDD394 pKa = 5.21DD395 pKa = 3.38IHH397 pKa = 6.17YY398 pKa = 7.46TGPSGIGPFNDD409 pKa = 3.4NNDD412 pKa = 3.67PSSASIGVYY421 pKa = 9.69TFDD424 pKa = 4.74ADD426 pKa = 3.77NKK428 pKa = 10.01PVYY431 pKa = 8.96QTGIEE436 pKa = 4.33GKK438 pKa = 10.17VEE440 pKa = 3.79
Molecular weight: 44.21 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0N0KW66|A0A0N0KW66_9MICO Transcriptional regulator OS=Frigoribacterium sp. RIT-PI-h OX=1690245 GN=AEQ27_12095 PE=4 SV=1
MM1 pKa = 7.94RR2 pKa = 11.84GPAPRR7 pKa = 11.84GRR9 pKa = 11.84RR10 pKa = 11.84GRR12 pKa = 11.84ARR14 pKa = 11.84PRR16 pKa = 11.84AASRR20 pKa = 11.84AAGNGAVLGRR30 pKa = 11.84TPRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84PRR37 pKa = 11.84RR38 pKa = 11.84PRR40 pKa = 11.84RR41 pKa = 11.84ASTRR45 pKa = 11.84RR46 pKa = 11.84DD47 pKa = 3.02RR48 pKa = 11.84SAPARR53 pKa = 11.84PSARR57 pKa = 11.84ARR59 pKa = 11.84TRR61 pKa = 11.84RR62 pKa = 11.84RR63 pKa = 11.84GRR65 pKa = 11.84PGGVRR70 pKa = 11.84RR71 pKa = 11.84RR72 pKa = 11.84AGRR75 pKa = 11.84SGPGRR80 pKa = 11.84GGAAAGGGWGSWSS93 pKa = 3.24
MM1 pKa = 7.94RR2 pKa = 11.84GPAPRR7 pKa = 11.84GRR9 pKa = 11.84RR10 pKa = 11.84GRR12 pKa = 11.84ARR14 pKa = 11.84PRR16 pKa = 11.84AASRR20 pKa = 11.84AAGNGAVLGRR30 pKa = 11.84TPRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84PRR37 pKa = 11.84RR38 pKa = 11.84PRR40 pKa = 11.84RR41 pKa = 11.84ASTRR45 pKa = 11.84RR46 pKa = 11.84DD47 pKa = 3.02RR48 pKa = 11.84SAPARR53 pKa = 11.84PSARR57 pKa = 11.84ARR59 pKa = 11.84TRR61 pKa = 11.84RR62 pKa = 11.84RR63 pKa = 11.84GRR65 pKa = 11.84PGGVRR70 pKa = 11.84RR71 pKa = 11.84RR72 pKa = 11.84AGRR75 pKa = 11.84SGPGRR80 pKa = 11.84GGAAAGGGWGSWSS93 pKa = 3.24
Molecular weight: 9.95 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
842116 |
38 |
3016 |
299.2 |
31.8 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.168 ± 0.055 | 0.45 ± 0.01 |
6.652 ± 0.045 | 5.194 ± 0.042 |
3.032 ± 0.029 | 9.321 ± 0.043 |
1.912 ± 0.026 | 3.687 ± 0.037 |
1.946 ± 0.032 | 9.927 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.627 ± 0.019 | 1.756 ± 0.021 |
5.564 ± 0.036 | 2.77 ± 0.031 |
7.254 ± 0.054 | 5.974 ± 0.033 |
6.523 ± 0.054 | 9.976 ± 0.058 |
1.389 ± 0.019 | 1.879 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
