
Dehalogenimonas formicexedens
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Chloroflexi; Dehalococcoidia; Dehalogenimonas
Average proteome isoelectric point is 6.67
Get precalculated fractions of proteins
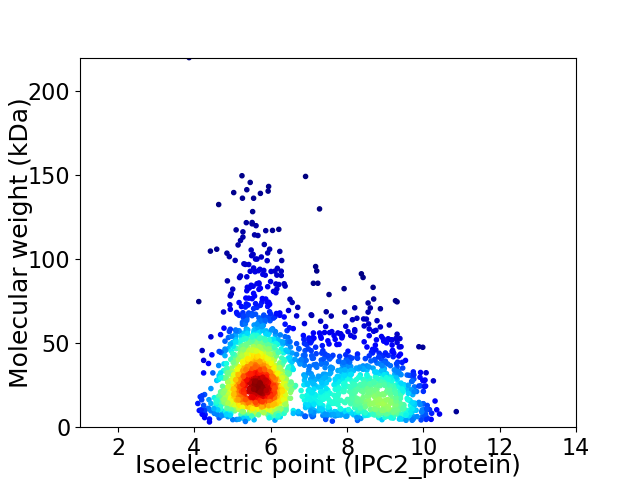
Virtual 2D-PAGE plot for 2117 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1P8F4T9|A0A1P8F4T9_9CHLR tRNA nucleotidyltransferase (CCA-adding enzyme) OS=Dehalogenimonas formicexedens OX=1839801 GN=cca PE=3 SV=1
MM1 pKa = 7.48TKK3 pKa = 10.24KK4 pKa = 8.73QTLFYY9 pKa = 10.22KK10 pKa = 10.35VAIIAAGLLATMAMLFAAIGPTKK33 pKa = 9.96TDD35 pKa = 3.2AAGGSVTQTSLGWRR49 pKa = 11.84NGPNAQVWGNGNYY62 pKa = 10.45GEE64 pKa = 4.41YY65 pKa = 10.26PEE67 pKa = 5.1GDD69 pKa = 3.16YY70 pKa = 10.11WQSQILITNGTSAAIDD86 pKa = 3.08ISGFGFILDD95 pKa = 4.73FYY97 pKa = 11.21NSGTVGVDD105 pKa = 2.99WVKK108 pKa = 11.11NLAWSMTPSAYY119 pKa = 9.72FVAGDD124 pKa = 4.35GPSDD128 pKa = 3.67RR129 pKa = 11.84YY130 pKa = 10.38PYY132 pKa = 8.64TGHH135 pKa = 6.76VAGDD139 pKa = 4.34DD140 pKa = 3.46IGLSDD145 pKa = 5.03WKK147 pKa = 9.91TGWTPFTPKK156 pKa = 10.02VINMPWDD163 pKa = 4.32PGTDD167 pKa = 3.16SYY169 pKa = 11.08ITTEE173 pKa = 4.15SQTAPAASHH182 pKa = 6.23FWGGFDD188 pKa = 4.18VNPPPTMLQPGEE200 pKa = 3.99SLAIYY205 pKa = 8.84FEE207 pKa = 4.14PHH209 pKa = 6.22LALTFIWSHH218 pKa = 5.11GAEE221 pKa = 4.02NLLPQIDD228 pKa = 4.0GVYY231 pKa = 8.96PGSGGTPTTFDD242 pKa = 3.95RR243 pKa = 11.84PWTAQYY249 pKa = 10.92NGAGFYY255 pKa = 10.21PGSSLHH261 pKa = 6.2GSIVYY266 pKa = 10.4SGLKK270 pKa = 9.05TFQLPSGAVVNGVISGYY287 pKa = 10.44KK288 pKa = 10.44YY289 pKa = 10.39NDD291 pKa = 3.45KK292 pKa = 10.85DD293 pKa = 3.63VSGTFTAGDD302 pKa = 3.47VGIPGWPIKK311 pKa = 10.57LYY313 pKa = 9.62ATVEE317 pKa = 4.27GVPITVSTVTDD328 pKa = 3.42SNGFYY333 pKa = 10.5QFTNLPYY340 pKa = 8.7GTYY343 pKa = 9.71SVKK346 pKa = 10.21EE347 pKa = 3.94APEE350 pKa = 4.19NGASIPAGYY359 pKa = 10.53EE360 pKa = 3.85NFVHH364 pKa = 6.7SFPANGTISGIVLTSPAPVSANNNFFNYY392 pKa = 8.3GTGKK396 pKa = 8.74LTVTKK401 pKa = 10.26SVVLGSVVNPAGISEE416 pKa = 4.6TFTIHH421 pKa = 3.52VTGPGGYY428 pKa = 9.17SHH430 pKa = 8.08DD431 pKa = 3.5ITFTLTNGVLQSPTSVTLEE450 pKa = 3.74GLIPGSYY457 pKa = 8.74TVSEE461 pKa = 4.72DD462 pKa = 3.43NPGAEE467 pKa = 4.02WTVTGGGSVTVNSGQTATSTVTNTFKK493 pKa = 10.76PGSLEE498 pKa = 3.57ITKK501 pKa = 10.69VIDD504 pKa = 3.45FNGAVNSASIDD515 pKa = 3.25EE516 pKa = 4.48TFTVKK521 pKa = 10.25VVGPSYY527 pKa = 10.14PGEE530 pKa = 4.35GTTHH534 pKa = 6.54QFVLDD539 pKa = 3.74NGAIVAPNPWLLTGLIPGDD558 pKa = 3.66YY559 pKa = 10.34TITEE563 pKa = 3.98VDD565 pKa = 4.91AGAEE569 pKa = 3.94WTEE572 pKa = 4.2TVPASAVAVSAGGVATADD590 pKa = 3.39VTNDD594 pKa = 3.48YY595 pKa = 11.16VPGSLEE601 pKa = 3.57ITKK604 pKa = 10.69VIDD607 pKa = 3.45FNGAVNSASIDD618 pKa = 3.25EE619 pKa = 4.48TFTVKK624 pKa = 10.25VVGPSYY630 pKa = 10.14PGEE633 pKa = 4.35GTTHH637 pKa = 6.54QFVLDD642 pKa = 3.74NGAIVAPNPWLLTGLIPGDD661 pKa = 3.66YY662 pKa = 10.34TITEE666 pKa = 3.98VDD668 pKa = 4.91AGAEE672 pKa = 3.94WTEE675 pKa = 4.2TVPASAVAVSAGVKK689 pKa = 8.85ATADD693 pKa = 3.44VTNDD697 pKa = 3.48YY698 pKa = 11.16VPGSLEE704 pKa = 3.42ITKK707 pKa = 10.31IIDD710 pKa = 3.12FNGAVNSASIDD721 pKa = 3.25EE722 pKa = 4.48TFTVKK727 pKa = 10.25VVGPSYY733 pKa = 10.14PGEE736 pKa = 4.35GTTHH740 pKa = 6.54QFVLDD745 pKa = 3.74NGAIVAPNPWLLTGLIPGDD764 pKa = 3.66YY765 pKa = 10.34TITEE769 pKa = 3.98VDD771 pKa = 4.91AGAEE775 pKa = 3.94WTEE778 pKa = 4.2TVPASAVAVSAGVKK792 pKa = 8.85ATADD796 pKa = 3.44VTNDD800 pKa = 3.48YY801 pKa = 11.16VPGSLEE807 pKa = 3.57ITKK810 pKa = 10.69VIDD813 pKa = 3.45FNGAVNSASIDD824 pKa = 3.25EE825 pKa = 4.48TFTVKK830 pKa = 10.25VVGPSYY836 pKa = 10.14PGEE839 pKa = 4.35GTTHH843 pKa = 6.54QFVLDD848 pKa = 3.74NGAIVAPNPWLLTGLIPGDD867 pKa = 3.66YY868 pKa = 10.34TITEE872 pKa = 3.98VDD874 pKa = 4.91AGAEE878 pKa = 3.94WTEE881 pKa = 4.2TVPASAVAVSAGGVATADD899 pKa = 3.39VTNDD903 pKa = 3.48YY904 pKa = 11.16VPGSLEE910 pKa = 3.57ITKK913 pKa = 10.69VIDD916 pKa = 3.45FNGAVNSASIDD927 pKa = 3.25EE928 pKa = 4.48TFTVKK933 pKa = 10.25VVGPSYY939 pKa = 10.14PGEE942 pKa = 4.35GTTHH946 pKa = 6.54QFVLDD951 pKa = 3.74NGAIVAPNPWLLTGLIPGDD970 pKa = 3.66YY971 pKa = 10.34TITEE975 pKa = 3.98VDD977 pKa = 4.91AGAEE981 pKa = 3.94WTEE984 pKa = 4.2TVPASAVAVSAGVKK998 pKa = 8.85ATADD1002 pKa = 3.44VTNDD1006 pKa = 3.48YY1007 pKa = 11.16VPGSLEE1013 pKa = 3.57ITKK1016 pKa = 10.69VIDD1019 pKa = 3.45FNGAVNSASIDD1030 pKa = 3.25EE1031 pKa = 4.48TFTVKK1036 pKa = 10.25VVGPSYY1042 pKa = 10.14PGEE1045 pKa = 4.35GTTHH1049 pKa = 6.54QFVLDD1054 pKa = 3.74NGAIVAPNPWLLTGLIPGDD1073 pKa = 3.66YY1074 pKa = 10.34TITEE1078 pKa = 3.98VDD1080 pKa = 4.91AGAEE1084 pKa = 3.94WTEE1087 pKa = 4.2TVPASAVAVSAGVKK1101 pKa = 8.85ATADD1105 pKa = 3.44VTNDD1109 pKa = 3.48YY1110 pKa = 11.16VPGSLEE1116 pKa = 3.42ITKK1119 pKa = 10.31IIDD1122 pKa = 3.12FNGAVNSASIDD1133 pKa = 3.25EE1134 pKa = 4.48TFTVKK1139 pKa = 10.25VVGPSYY1145 pKa = 10.14PGEE1148 pKa = 4.35GTTHH1152 pKa = 6.54QFVLDD1157 pKa = 3.74NGAIVAPNPWLLTGLIPGDD1176 pKa = 3.66YY1177 pKa = 10.34TITEE1181 pKa = 3.98VDD1183 pKa = 4.91AGAEE1187 pKa = 3.94WTEE1190 pKa = 4.2TVPASAVAVSAGVKK1204 pKa = 8.85ATADD1208 pKa = 3.44VTNDD1212 pKa = 3.48YY1213 pKa = 11.16VPGSLEE1219 pKa = 3.57ITKK1222 pKa = 10.69VIDD1225 pKa = 3.45FNGAVNSASIDD1236 pKa = 3.25EE1237 pKa = 4.48TFTVKK1242 pKa = 10.25VVGPSYY1248 pKa = 10.14PGEE1251 pKa = 4.35GTTHH1255 pKa = 6.54QFVLDD1260 pKa = 3.74NGAIVAPNPWLLTGLIPGDD1279 pKa = 3.66YY1280 pKa = 10.34TITEE1284 pKa = 3.98VDD1286 pKa = 4.91AGAEE1290 pKa = 3.94WTEE1293 pKa = 4.2TVPASAVAVSAGGVATADD1311 pKa = 3.39VTNDD1315 pKa = 3.48YY1316 pKa = 11.16VPGSLEE1322 pKa = 3.57ITKK1325 pKa = 10.69VIDD1328 pKa = 3.45FNGAVNSASIDD1339 pKa = 3.25EE1340 pKa = 4.48TFTVKK1345 pKa = 10.25VVGPSYY1351 pKa = 10.14PGEE1354 pKa = 4.35GTTHH1358 pKa = 6.54QFVLDD1363 pKa = 3.74NGAIVAPNPWLLTGLIPGDD1382 pKa = 3.66YY1383 pKa = 10.34TITEE1387 pKa = 3.98VDD1389 pKa = 4.91AGAEE1393 pKa = 3.94WTEE1396 pKa = 4.2TVPASAVAVSAGVKK1410 pKa = 8.85ATADD1414 pKa = 3.44VTNDD1418 pKa = 3.48YY1419 pKa = 11.16VPGSLEE1425 pKa = 3.42ITKK1428 pKa = 10.31IIDD1431 pKa = 3.12FNGAVNSASIDD1442 pKa = 3.25EE1443 pKa = 4.48TFTVKK1448 pKa = 10.25VVGPSYY1454 pKa = 10.14PGEE1457 pKa = 4.35GTTHH1461 pKa = 6.54QFVLDD1466 pKa = 3.74NGAIVAPNPWLLTGLIPGDD1485 pKa = 3.66YY1486 pKa = 10.34TITEE1490 pKa = 3.98VDD1492 pKa = 4.91AGAEE1496 pKa = 3.94WTEE1499 pKa = 4.2TVPASAVAVSAGGVATADD1517 pKa = 3.39VTNDD1521 pKa = 3.48YY1522 pKa = 11.16VPGSLEE1528 pKa = 3.57ITKK1531 pKa = 10.69VIDD1534 pKa = 3.45FNGAVNSASIDD1545 pKa = 3.25EE1546 pKa = 4.48TFTVKK1551 pKa = 10.25VVGPSYY1557 pKa = 10.14PGEE1560 pKa = 4.35GTTHH1564 pKa = 6.54QFVLDD1569 pKa = 3.74NGAIVAPNPWLLTGLIPGDD1588 pKa = 3.66YY1589 pKa = 10.34TITEE1593 pKa = 3.98VDD1595 pKa = 4.91AGAEE1599 pKa = 3.94WTEE1602 pKa = 4.2TVPASAVAVSAGVKK1616 pKa = 8.85ATADD1620 pKa = 3.44VTNDD1624 pKa = 3.48YY1625 pKa = 11.16VPGSLEE1631 pKa = 3.42ITKK1634 pKa = 10.31IIDD1637 pKa = 3.12FNGAVNSASIDD1648 pKa = 3.25EE1649 pKa = 4.48TFTVKK1654 pKa = 10.25VVGPSYY1660 pKa = 10.14PGEE1663 pKa = 4.35GTTHH1667 pKa = 6.54QFVLDD1672 pKa = 3.74NGAIVAPNPWLLTGLIPGDD1691 pKa = 3.66YY1692 pKa = 10.34TITEE1696 pKa = 3.98VDD1698 pKa = 4.91AGAEE1702 pKa = 3.94WTEE1705 pKa = 4.2TVPASAVAVSAGVKK1719 pKa = 8.85ATADD1723 pKa = 3.47VTNDD1727 pKa = 3.51FVPGSLTITKK1737 pKa = 9.72VVVLGAYY1744 pKa = 8.79PFASTTTLDD1753 pKa = 3.44FTVKK1757 pKa = 8.92VTGPSYY1763 pKa = 10.21PGPDD1767 pKa = 3.08GTTLTFNLVNGVISGAQTLDD1787 pKa = 3.21NLIPGVYY1794 pKa = 8.82TVTEE1798 pKa = 4.15TDD1800 pKa = 3.74PGVAWTVTNLTGNVTVDD1817 pKa = 3.61PGTPTATRR1825 pKa = 11.84TITNTLKK1832 pKa = 10.63QPNTTVTIGSDD1843 pKa = 2.83TWEE1846 pKa = 4.28TFPGGNVTITVTEE1859 pKa = 4.38EE1860 pKa = 3.72NTGDD1864 pKa = 3.58VPLSNVSVVVTQNDD1878 pKa = 4.2GSPLTLSAPPTSGDD1892 pKa = 3.52ANTDD1896 pKa = 3.8GILDD1900 pKa = 3.94TDD1902 pKa = 4.25EE1903 pKa = 3.95TWTWVYY1909 pKa = 9.69LTTISVDD1916 pKa = 3.35TTFQATGHH1924 pKa = 5.69GTDD1927 pKa = 4.09PLGNDD1932 pKa = 2.75ITVPNYY1938 pKa = 7.96PTEE1941 pKa = 4.0QDD1943 pKa = 3.51DD1944 pKa = 4.48VFVEE1948 pKa = 4.45VNGATRR1954 pKa = 11.84TIGFWKK1960 pKa = 7.6THH1962 pKa = 4.69WDD1964 pKa = 3.48FTEE1967 pKa = 4.71HH1968 pKa = 6.5VFTDD1972 pKa = 3.74PTGLNSNINLGTWGGKK1988 pKa = 6.03TWTITTMEE1996 pKa = 4.17QLMGLLWANTAKK2008 pKa = 10.78NADD2011 pKa = 3.86EE2012 pKa = 4.39TSSSRR2017 pKa = 11.84LTIDD2021 pKa = 2.86QAKK2024 pKa = 9.6IHH2026 pKa = 5.92TAQQALAAILNDD2038 pKa = 3.38AMAGGAPLPVTLTDD2052 pKa = 3.06IVGVLEE2058 pKa = 4.57GNSIGQVRR2066 pKa = 11.84SLGSTLDD2073 pKa = 3.41AYY2075 pKa = 11.42NNSGDD2080 pKa = 4.58NIALDD2085 pKa = 3.88PSLPPTKK2092 pKa = 10.3KK2093 pKa = 10.48GDD2095 pKa = 3.39EE2096 pKa = 4.37GKK2098 pKa = 9.83PKK2100 pKa = 10.63DD2101 pKa = 4.07LADD2104 pKa = 3.93IPWANTTPEE2113 pKa = 3.88APKK2116 pKa = 10.63GKK2118 pKa = 10.09KK2119 pKa = 9.15
MM1 pKa = 7.48TKK3 pKa = 10.24KK4 pKa = 8.73QTLFYY9 pKa = 10.22KK10 pKa = 10.35VAIIAAGLLATMAMLFAAIGPTKK33 pKa = 9.96TDD35 pKa = 3.2AAGGSVTQTSLGWRR49 pKa = 11.84NGPNAQVWGNGNYY62 pKa = 10.45GEE64 pKa = 4.41YY65 pKa = 10.26PEE67 pKa = 5.1GDD69 pKa = 3.16YY70 pKa = 10.11WQSQILITNGTSAAIDD86 pKa = 3.08ISGFGFILDD95 pKa = 4.73FYY97 pKa = 11.21NSGTVGVDD105 pKa = 2.99WVKK108 pKa = 11.11NLAWSMTPSAYY119 pKa = 9.72FVAGDD124 pKa = 4.35GPSDD128 pKa = 3.67RR129 pKa = 11.84YY130 pKa = 10.38PYY132 pKa = 8.64TGHH135 pKa = 6.76VAGDD139 pKa = 4.34DD140 pKa = 3.46IGLSDD145 pKa = 5.03WKK147 pKa = 9.91TGWTPFTPKK156 pKa = 10.02VINMPWDD163 pKa = 4.32PGTDD167 pKa = 3.16SYY169 pKa = 11.08ITTEE173 pKa = 4.15SQTAPAASHH182 pKa = 6.23FWGGFDD188 pKa = 4.18VNPPPTMLQPGEE200 pKa = 3.99SLAIYY205 pKa = 8.84FEE207 pKa = 4.14PHH209 pKa = 6.22LALTFIWSHH218 pKa = 5.11GAEE221 pKa = 4.02NLLPQIDD228 pKa = 4.0GVYY231 pKa = 8.96PGSGGTPTTFDD242 pKa = 3.95RR243 pKa = 11.84PWTAQYY249 pKa = 10.92NGAGFYY255 pKa = 10.21PGSSLHH261 pKa = 6.2GSIVYY266 pKa = 10.4SGLKK270 pKa = 9.05TFQLPSGAVVNGVISGYY287 pKa = 10.44KK288 pKa = 10.44YY289 pKa = 10.39NDD291 pKa = 3.45KK292 pKa = 10.85DD293 pKa = 3.63VSGTFTAGDD302 pKa = 3.47VGIPGWPIKK311 pKa = 10.57LYY313 pKa = 9.62ATVEE317 pKa = 4.27GVPITVSTVTDD328 pKa = 3.42SNGFYY333 pKa = 10.5QFTNLPYY340 pKa = 8.7GTYY343 pKa = 9.71SVKK346 pKa = 10.21EE347 pKa = 3.94APEE350 pKa = 4.19NGASIPAGYY359 pKa = 10.53EE360 pKa = 3.85NFVHH364 pKa = 6.7SFPANGTISGIVLTSPAPVSANNNFFNYY392 pKa = 8.3GTGKK396 pKa = 8.74LTVTKK401 pKa = 10.26SVVLGSVVNPAGISEE416 pKa = 4.6TFTIHH421 pKa = 3.52VTGPGGYY428 pKa = 9.17SHH430 pKa = 8.08DD431 pKa = 3.5ITFTLTNGVLQSPTSVTLEE450 pKa = 3.74GLIPGSYY457 pKa = 8.74TVSEE461 pKa = 4.72DD462 pKa = 3.43NPGAEE467 pKa = 4.02WTVTGGGSVTVNSGQTATSTVTNTFKK493 pKa = 10.76PGSLEE498 pKa = 3.57ITKK501 pKa = 10.69VIDD504 pKa = 3.45FNGAVNSASIDD515 pKa = 3.25EE516 pKa = 4.48TFTVKK521 pKa = 10.25VVGPSYY527 pKa = 10.14PGEE530 pKa = 4.35GTTHH534 pKa = 6.54QFVLDD539 pKa = 3.74NGAIVAPNPWLLTGLIPGDD558 pKa = 3.66YY559 pKa = 10.34TITEE563 pKa = 3.98VDD565 pKa = 4.91AGAEE569 pKa = 3.94WTEE572 pKa = 4.2TVPASAVAVSAGGVATADD590 pKa = 3.39VTNDD594 pKa = 3.48YY595 pKa = 11.16VPGSLEE601 pKa = 3.57ITKK604 pKa = 10.69VIDD607 pKa = 3.45FNGAVNSASIDD618 pKa = 3.25EE619 pKa = 4.48TFTVKK624 pKa = 10.25VVGPSYY630 pKa = 10.14PGEE633 pKa = 4.35GTTHH637 pKa = 6.54QFVLDD642 pKa = 3.74NGAIVAPNPWLLTGLIPGDD661 pKa = 3.66YY662 pKa = 10.34TITEE666 pKa = 3.98VDD668 pKa = 4.91AGAEE672 pKa = 3.94WTEE675 pKa = 4.2TVPASAVAVSAGVKK689 pKa = 8.85ATADD693 pKa = 3.44VTNDD697 pKa = 3.48YY698 pKa = 11.16VPGSLEE704 pKa = 3.42ITKK707 pKa = 10.31IIDD710 pKa = 3.12FNGAVNSASIDD721 pKa = 3.25EE722 pKa = 4.48TFTVKK727 pKa = 10.25VVGPSYY733 pKa = 10.14PGEE736 pKa = 4.35GTTHH740 pKa = 6.54QFVLDD745 pKa = 3.74NGAIVAPNPWLLTGLIPGDD764 pKa = 3.66YY765 pKa = 10.34TITEE769 pKa = 3.98VDD771 pKa = 4.91AGAEE775 pKa = 3.94WTEE778 pKa = 4.2TVPASAVAVSAGVKK792 pKa = 8.85ATADD796 pKa = 3.44VTNDD800 pKa = 3.48YY801 pKa = 11.16VPGSLEE807 pKa = 3.57ITKK810 pKa = 10.69VIDD813 pKa = 3.45FNGAVNSASIDD824 pKa = 3.25EE825 pKa = 4.48TFTVKK830 pKa = 10.25VVGPSYY836 pKa = 10.14PGEE839 pKa = 4.35GTTHH843 pKa = 6.54QFVLDD848 pKa = 3.74NGAIVAPNPWLLTGLIPGDD867 pKa = 3.66YY868 pKa = 10.34TITEE872 pKa = 3.98VDD874 pKa = 4.91AGAEE878 pKa = 3.94WTEE881 pKa = 4.2TVPASAVAVSAGGVATADD899 pKa = 3.39VTNDD903 pKa = 3.48YY904 pKa = 11.16VPGSLEE910 pKa = 3.57ITKK913 pKa = 10.69VIDD916 pKa = 3.45FNGAVNSASIDD927 pKa = 3.25EE928 pKa = 4.48TFTVKK933 pKa = 10.25VVGPSYY939 pKa = 10.14PGEE942 pKa = 4.35GTTHH946 pKa = 6.54QFVLDD951 pKa = 3.74NGAIVAPNPWLLTGLIPGDD970 pKa = 3.66YY971 pKa = 10.34TITEE975 pKa = 3.98VDD977 pKa = 4.91AGAEE981 pKa = 3.94WTEE984 pKa = 4.2TVPASAVAVSAGVKK998 pKa = 8.85ATADD1002 pKa = 3.44VTNDD1006 pKa = 3.48YY1007 pKa = 11.16VPGSLEE1013 pKa = 3.57ITKK1016 pKa = 10.69VIDD1019 pKa = 3.45FNGAVNSASIDD1030 pKa = 3.25EE1031 pKa = 4.48TFTVKK1036 pKa = 10.25VVGPSYY1042 pKa = 10.14PGEE1045 pKa = 4.35GTTHH1049 pKa = 6.54QFVLDD1054 pKa = 3.74NGAIVAPNPWLLTGLIPGDD1073 pKa = 3.66YY1074 pKa = 10.34TITEE1078 pKa = 3.98VDD1080 pKa = 4.91AGAEE1084 pKa = 3.94WTEE1087 pKa = 4.2TVPASAVAVSAGVKK1101 pKa = 8.85ATADD1105 pKa = 3.44VTNDD1109 pKa = 3.48YY1110 pKa = 11.16VPGSLEE1116 pKa = 3.42ITKK1119 pKa = 10.31IIDD1122 pKa = 3.12FNGAVNSASIDD1133 pKa = 3.25EE1134 pKa = 4.48TFTVKK1139 pKa = 10.25VVGPSYY1145 pKa = 10.14PGEE1148 pKa = 4.35GTTHH1152 pKa = 6.54QFVLDD1157 pKa = 3.74NGAIVAPNPWLLTGLIPGDD1176 pKa = 3.66YY1177 pKa = 10.34TITEE1181 pKa = 3.98VDD1183 pKa = 4.91AGAEE1187 pKa = 3.94WTEE1190 pKa = 4.2TVPASAVAVSAGVKK1204 pKa = 8.85ATADD1208 pKa = 3.44VTNDD1212 pKa = 3.48YY1213 pKa = 11.16VPGSLEE1219 pKa = 3.57ITKK1222 pKa = 10.69VIDD1225 pKa = 3.45FNGAVNSASIDD1236 pKa = 3.25EE1237 pKa = 4.48TFTVKK1242 pKa = 10.25VVGPSYY1248 pKa = 10.14PGEE1251 pKa = 4.35GTTHH1255 pKa = 6.54QFVLDD1260 pKa = 3.74NGAIVAPNPWLLTGLIPGDD1279 pKa = 3.66YY1280 pKa = 10.34TITEE1284 pKa = 3.98VDD1286 pKa = 4.91AGAEE1290 pKa = 3.94WTEE1293 pKa = 4.2TVPASAVAVSAGGVATADD1311 pKa = 3.39VTNDD1315 pKa = 3.48YY1316 pKa = 11.16VPGSLEE1322 pKa = 3.57ITKK1325 pKa = 10.69VIDD1328 pKa = 3.45FNGAVNSASIDD1339 pKa = 3.25EE1340 pKa = 4.48TFTVKK1345 pKa = 10.25VVGPSYY1351 pKa = 10.14PGEE1354 pKa = 4.35GTTHH1358 pKa = 6.54QFVLDD1363 pKa = 3.74NGAIVAPNPWLLTGLIPGDD1382 pKa = 3.66YY1383 pKa = 10.34TITEE1387 pKa = 3.98VDD1389 pKa = 4.91AGAEE1393 pKa = 3.94WTEE1396 pKa = 4.2TVPASAVAVSAGVKK1410 pKa = 8.85ATADD1414 pKa = 3.44VTNDD1418 pKa = 3.48YY1419 pKa = 11.16VPGSLEE1425 pKa = 3.42ITKK1428 pKa = 10.31IIDD1431 pKa = 3.12FNGAVNSASIDD1442 pKa = 3.25EE1443 pKa = 4.48TFTVKK1448 pKa = 10.25VVGPSYY1454 pKa = 10.14PGEE1457 pKa = 4.35GTTHH1461 pKa = 6.54QFVLDD1466 pKa = 3.74NGAIVAPNPWLLTGLIPGDD1485 pKa = 3.66YY1486 pKa = 10.34TITEE1490 pKa = 3.98VDD1492 pKa = 4.91AGAEE1496 pKa = 3.94WTEE1499 pKa = 4.2TVPASAVAVSAGGVATADD1517 pKa = 3.39VTNDD1521 pKa = 3.48YY1522 pKa = 11.16VPGSLEE1528 pKa = 3.57ITKK1531 pKa = 10.69VIDD1534 pKa = 3.45FNGAVNSASIDD1545 pKa = 3.25EE1546 pKa = 4.48TFTVKK1551 pKa = 10.25VVGPSYY1557 pKa = 10.14PGEE1560 pKa = 4.35GTTHH1564 pKa = 6.54QFVLDD1569 pKa = 3.74NGAIVAPNPWLLTGLIPGDD1588 pKa = 3.66YY1589 pKa = 10.34TITEE1593 pKa = 3.98VDD1595 pKa = 4.91AGAEE1599 pKa = 3.94WTEE1602 pKa = 4.2TVPASAVAVSAGVKK1616 pKa = 8.85ATADD1620 pKa = 3.44VTNDD1624 pKa = 3.48YY1625 pKa = 11.16VPGSLEE1631 pKa = 3.42ITKK1634 pKa = 10.31IIDD1637 pKa = 3.12FNGAVNSASIDD1648 pKa = 3.25EE1649 pKa = 4.48TFTVKK1654 pKa = 10.25VVGPSYY1660 pKa = 10.14PGEE1663 pKa = 4.35GTTHH1667 pKa = 6.54QFVLDD1672 pKa = 3.74NGAIVAPNPWLLTGLIPGDD1691 pKa = 3.66YY1692 pKa = 10.34TITEE1696 pKa = 3.98VDD1698 pKa = 4.91AGAEE1702 pKa = 3.94WTEE1705 pKa = 4.2TVPASAVAVSAGVKK1719 pKa = 8.85ATADD1723 pKa = 3.47VTNDD1727 pKa = 3.51FVPGSLTITKK1737 pKa = 9.72VVVLGAYY1744 pKa = 8.79PFASTTTLDD1753 pKa = 3.44FTVKK1757 pKa = 8.92VTGPSYY1763 pKa = 10.21PGPDD1767 pKa = 3.08GTTLTFNLVNGVISGAQTLDD1787 pKa = 3.21NLIPGVYY1794 pKa = 8.82TVTEE1798 pKa = 4.15TDD1800 pKa = 3.74PGVAWTVTNLTGNVTVDD1817 pKa = 3.61PGTPTATRR1825 pKa = 11.84TITNTLKK1832 pKa = 10.63QPNTTVTIGSDD1843 pKa = 2.83TWEE1846 pKa = 4.28TFPGGNVTITVTEE1859 pKa = 4.38EE1860 pKa = 3.72NTGDD1864 pKa = 3.58VPLSNVSVVVTQNDD1878 pKa = 4.2GSPLTLSAPPTSGDD1892 pKa = 3.52ANTDD1896 pKa = 3.8GILDD1900 pKa = 3.94TDD1902 pKa = 4.25EE1903 pKa = 3.95TWTWVYY1909 pKa = 9.69LTTISVDD1916 pKa = 3.35TTFQATGHH1924 pKa = 5.69GTDD1927 pKa = 4.09PLGNDD1932 pKa = 2.75ITVPNYY1938 pKa = 7.96PTEE1941 pKa = 4.0QDD1943 pKa = 3.51DD1944 pKa = 4.48VFVEE1948 pKa = 4.45VNGATRR1954 pKa = 11.84TIGFWKK1960 pKa = 7.6THH1962 pKa = 4.69WDD1964 pKa = 3.48FTEE1967 pKa = 4.71HH1968 pKa = 6.5VFTDD1972 pKa = 3.74PTGLNSNINLGTWGGKK1988 pKa = 6.03TWTITTMEE1996 pKa = 4.17QLMGLLWANTAKK2008 pKa = 10.78NADD2011 pKa = 3.86EE2012 pKa = 4.39TSSSRR2017 pKa = 11.84LTIDD2021 pKa = 2.86QAKK2024 pKa = 9.6IHH2026 pKa = 5.92TAQQALAAILNDD2038 pKa = 3.38AMAGGAPLPVTLTDD2052 pKa = 3.06IVGVLEE2058 pKa = 4.57GNSIGQVRR2066 pKa = 11.84SLGSTLDD2073 pKa = 3.41AYY2075 pKa = 11.42NNSGDD2080 pKa = 4.58NIALDD2085 pKa = 3.88PSLPPTKK2092 pKa = 10.3KK2093 pKa = 10.48GDD2095 pKa = 3.39EE2096 pKa = 4.37GKK2098 pKa = 9.83PKK2100 pKa = 10.63DD2101 pKa = 4.07LADD2104 pKa = 3.93IPWANTTPEE2113 pKa = 3.88APKK2116 pKa = 10.63GKK2118 pKa = 10.09KK2119 pKa = 9.15
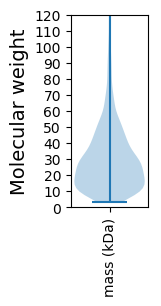
Molecular weight: 219.59 kDa
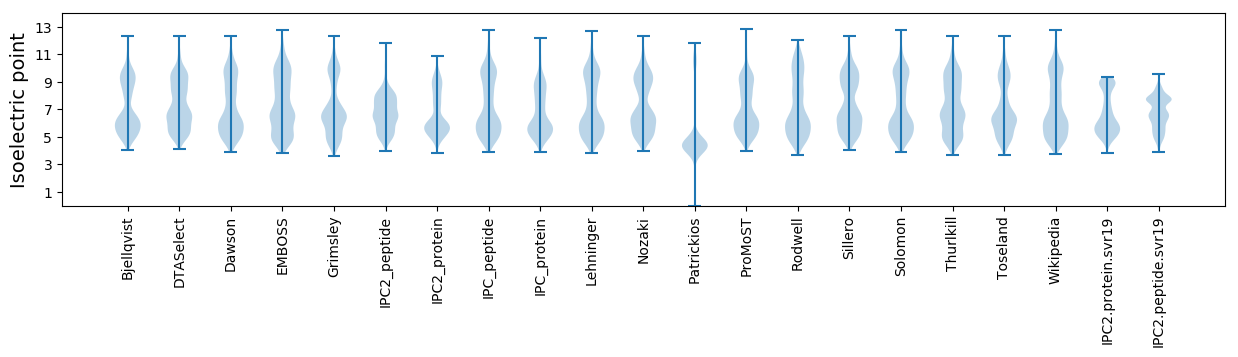
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1P8F9V0|A0A1P8F9V0_9CHLR 4Fe-4S binding domain-containing protein OS=Dehalogenimonas formicexedens OX=1839801 GN=Dform_01895 PE=4 SV=1
MM1 pKa = 7.87PDD3 pKa = 2.77IMSRR7 pKa = 11.84AIIQSGGGGGYY18 pKa = 9.41IEE20 pKa = 4.81GGRR23 pKa = 11.84VKK25 pKa = 10.56NPPLQCSDD33 pKa = 3.64SQTVAAGLQTSADD46 pKa = 3.33PSLRR50 pKa = 11.84QNNEE54 pKa = 3.35RR55 pKa = 11.84CVRR58 pKa = 11.84GWNGGGRR65 pKa = 11.84VKK67 pKa = 10.65NPPLQCPGAKK77 pKa = 9.51PGQPVSRR84 pKa = 11.84PAQIPVFAKK93 pKa = 8.57ITRR96 pKa = 11.84GAFDD100 pKa = 4.89DD101 pKa = 4.28EE102 pKa = 4.79PGEE105 pKa = 4.69GGLKK109 pKa = 8.37TRR111 pKa = 11.84PYY113 pKa = 10.03DD114 pKa = 3.33TGRR117 pKa = 11.84KK118 pKa = 8.31PGAPGVLWRR127 pKa = 11.84SRR129 pKa = 11.84RR130 pKa = 11.84RR131 pKa = 11.84RR132 pKa = 11.84DD133 pKa = 2.96SGLRR137 pKa = 11.84RR138 pKa = 11.84NDD140 pKa = 2.9KK141 pKa = 10.69RR142 pKa = 11.84HH143 pKa = 6.25LKK145 pKa = 10.76DD146 pKa = 3.04EE147 pKa = 4.29TGRR150 pKa = 11.84AGG152 pKa = 3.39
MM1 pKa = 7.87PDD3 pKa = 2.77IMSRR7 pKa = 11.84AIIQSGGGGGYY18 pKa = 9.41IEE20 pKa = 4.81GGRR23 pKa = 11.84VKK25 pKa = 10.56NPPLQCSDD33 pKa = 3.64SQTVAAGLQTSADD46 pKa = 3.33PSLRR50 pKa = 11.84QNNEE54 pKa = 3.35RR55 pKa = 11.84CVRR58 pKa = 11.84GWNGGGRR65 pKa = 11.84VKK67 pKa = 10.65NPPLQCPGAKK77 pKa = 9.51PGQPVSRR84 pKa = 11.84PAQIPVFAKK93 pKa = 8.57ITRR96 pKa = 11.84GAFDD100 pKa = 4.89DD101 pKa = 4.28EE102 pKa = 4.79PGEE105 pKa = 4.69GGLKK109 pKa = 8.37TRR111 pKa = 11.84PYY113 pKa = 10.03DD114 pKa = 3.33TGRR117 pKa = 11.84KK118 pKa = 8.31PGAPGVLWRR127 pKa = 11.84SRR129 pKa = 11.84RR130 pKa = 11.84RR131 pKa = 11.84RR132 pKa = 11.84DD133 pKa = 2.96SGLRR137 pKa = 11.84RR138 pKa = 11.84NDD140 pKa = 2.9KK141 pKa = 10.69RR142 pKa = 11.84HH143 pKa = 6.25LKK145 pKa = 10.76DD146 pKa = 3.04EE147 pKa = 4.29TGRR150 pKa = 11.84AGG152 pKa = 3.39
Molecular weight: 16.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
607201 |
29 |
2119 |
286.8 |
31.5 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.503 ± 0.06 | 1.161 ± 0.023 |
5.135 ± 0.034 | 6.152 ± 0.062 |
4.038 ± 0.04 | 8.002 ± 0.056 |
1.862 ± 0.021 | 6.676 ± 0.047 |
5.169 ± 0.052 | 9.881 ± 0.063 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.444 ± 0.028 | 3.458 ± 0.031 |
4.792 ± 0.038 | 3.059 ± 0.035 |
5.67 ± 0.057 | 6.145 ± 0.039 |
5.581 ± 0.061 | 7.328 ± 0.048 |
1.168 ± 0.021 | 2.777 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
