
Sphingomonas indica
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; Sphingomonadaceae; Allosphingosinicella
Average proteome isoelectric point is 6.48
Get precalculated fractions of proteins
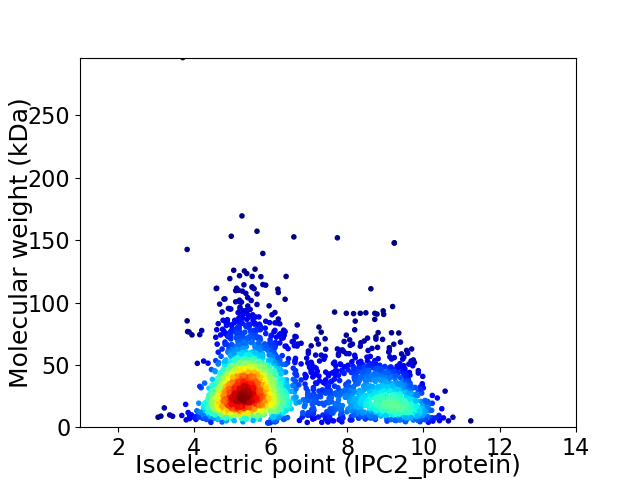
Virtual 2D-PAGE plot for 2739 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
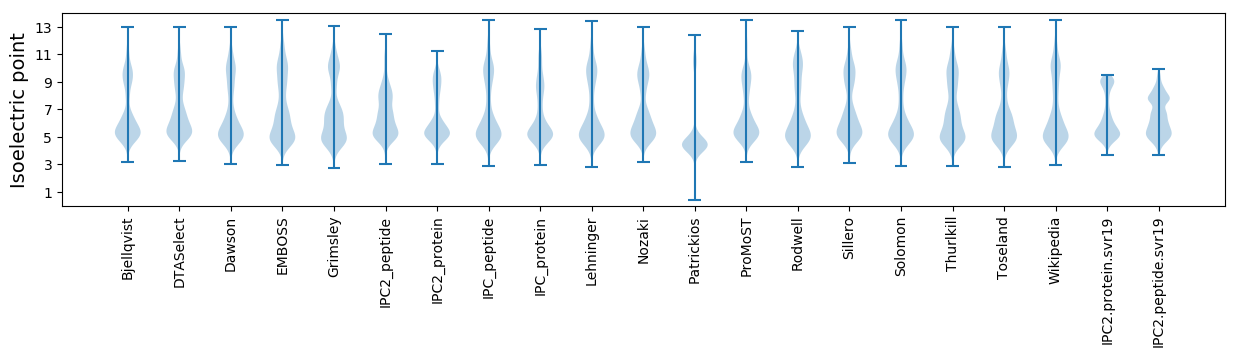
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1X7GD15|A0A1X7GD15_9SPHN 50S ribosomal protein L4 OS=Sphingomonas indica OX=941907 GN=rplD PE=3 SV=1
MM1 pKa = 7.43VGTLKK6 pKa = 10.72DD7 pKa = 3.26IEE9 pKa = 4.43FEE11 pKa = 4.26GDD13 pKa = 2.88RR14 pKa = 11.84STRR17 pKa = 11.84DD18 pKa = 3.3LNNEE22 pKa = 3.57YY23 pKa = 11.0SDD25 pKa = 4.57AYY27 pKa = 9.47VAAVRR32 pKa = 11.84ASQANVDD39 pKa = 4.06GQGGTIRR46 pKa = 11.84GKK48 pKa = 9.58PVFTKK53 pKa = 10.91EE54 pKa = 3.49EE55 pKa = 3.57VAYY58 pKa = 9.95YY59 pKa = 10.51LNRR62 pKa = 11.84GDD64 pKa = 4.38GIIGGFEE71 pKa = 4.52SGANWDD77 pKa = 3.83GAQGYY82 pKa = 10.75ANNQYY87 pKa = 10.46YY88 pKa = 9.07WLPVTKK94 pKa = 10.68AMGGEE99 pKa = 3.87AGLPSEE105 pKa = 5.11GGPGATGPLTTLTFGFYY122 pKa = 7.63EE123 pKa = 4.4TLASLPEE130 pKa = 4.21PYY132 pKa = 10.24VYY134 pKa = 9.03RR135 pKa = 11.84TPAGNLALGLSVAQGFSPFTAAQRR159 pKa = 11.84TATRR163 pKa = 11.84EE164 pKa = 4.08AIGLWDD170 pKa = 6.15DD171 pKa = 3.95IIKK174 pKa = 10.87VNFQEE179 pKa = 4.84TSFQQGDD186 pKa = 4.26LNFMNTTTGPAQAAAYY202 pKa = 9.78LPYY205 pKa = 10.39DD206 pKa = 3.76YY207 pKa = 11.2GSEE210 pKa = 3.78EE211 pKa = 3.96GFKK214 pKa = 10.88NFEE217 pKa = 4.31GDD219 pKa = 3.37DD220 pKa = 3.46VSYY223 pKa = 11.51YY224 pKa = 10.52EE225 pKa = 3.74IAGDD229 pKa = 3.72VFVNPNQASNFQFLPGQYY247 pKa = 10.83GLNTLVHH254 pKa = 6.98EE255 pKa = 4.85IGHH258 pKa = 6.83AIGLEE263 pKa = 4.03HH264 pKa = 7.18PGAYY268 pKa = 10.25NFGPGFDD275 pKa = 3.33VTYY278 pKa = 11.0DD279 pKa = 3.31NGAEE283 pKa = 4.15YY284 pKa = 10.88YY285 pKa = 9.8QDD287 pKa = 2.89SRR289 pKa = 11.84MYY291 pKa = 10.86SIMSYY296 pKa = 9.7WDD298 pKa = 3.44AEE300 pKa = 4.43EE301 pKa = 3.96TGAAHH306 pKa = 6.68VNWEE310 pKa = 4.07NLTYY314 pKa = 10.38SYY316 pKa = 11.38NSTPMIHH323 pKa = 7.7DD324 pKa = 3.66ILAAQRR330 pKa = 11.84IYY332 pKa = 10.86GVEE335 pKa = 3.91EE336 pKa = 3.59NTRR339 pKa = 11.84TGNTTYY345 pKa = 11.09GFNSNAGKK353 pKa = 10.52DD354 pKa = 3.45MYY356 pKa = 11.4DD357 pKa = 3.23FTKK360 pKa = 10.8NPNPVISIYY369 pKa = 10.65DD370 pKa = 3.29AGGNDD375 pKa = 3.59TLDD378 pKa = 3.31LSGYY382 pKa = 6.28NTRR385 pKa = 11.84SFIDD389 pKa = 3.82LNPGAFSSAGGFYY402 pKa = 10.64SEE404 pKa = 6.3DD405 pKa = 3.43IPTLDD410 pKa = 4.11EE411 pKa = 4.14INARR415 pKa = 11.84RR416 pKa = 11.84AAAGLAPRR424 pKa = 11.84TQATYY429 pKa = 11.23DD430 pKa = 3.5LYY432 pKa = 10.77IEE434 pKa = 5.14LFGATYY440 pKa = 10.46TDD442 pKa = 3.24GLMRR446 pKa = 11.84DD447 pKa = 4.41NISIAYY453 pKa = 9.83NSIIEE458 pKa = 4.13NAVGGGGDD466 pKa = 3.67DD467 pKa = 3.62TIVANGAANRR477 pKa = 11.84IDD479 pKa = 3.83GGAGNDD485 pKa = 2.92IVSYY489 pKa = 8.05QTSGTAVSVNLANGLVSGGAAGDD512 pKa = 3.56VLISIEE518 pKa = 4.13GVTGSRR524 pKa = 11.84HH525 pKa = 5.53NDD527 pKa = 2.86VLTGSDD533 pKa = 3.75GNNVLNGGAGNDD545 pKa = 3.42IIYY548 pKa = 10.2GRR550 pKa = 11.84NGDD553 pKa = 3.77DD554 pKa = 4.28TIDD557 pKa = 3.86GGDD560 pKa = 3.88GVDD563 pKa = 3.81QLFGDD568 pKa = 4.82GGNDD572 pKa = 3.81TIRR575 pKa = 11.84GGDD578 pKa = 3.68GNDD581 pKa = 3.69TIMGGAGDD589 pKa = 5.58DD590 pKa = 3.92ILFGDD595 pKa = 4.76AGNDD599 pKa = 3.48TLYY602 pKa = 11.38GDD604 pKa = 4.75AGNDD608 pKa = 3.57VLDD611 pKa = 4.97GGAGDD616 pKa = 4.06DD617 pKa = 4.12KK618 pKa = 11.52LYY620 pKa = 11.25GGAGADD626 pKa = 3.3TFRR629 pKa = 11.84FSDD632 pKa = 3.44LGGYY636 pKa = 7.53DD637 pKa = 3.41TIVDD641 pKa = 4.24FRR643 pKa = 11.84RR644 pKa = 11.84GQGDD648 pKa = 3.92KK649 pKa = 10.48IDD651 pKa = 3.94LTGIDD656 pKa = 3.78PNINVDD662 pKa = 3.62AVNEE666 pKa = 3.94QFNFIGNAAFSGTAGEE682 pKa = 4.54LRR684 pKa = 11.84TFVDD688 pKa = 3.4RR689 pKa = 11.84GSTIIAGDD697 pKa = 3.61VNGDD701 pKa = 3.39GVADD705 pKa = 3.93FQINVGATAIRR716 pKa = 11.84ADD718 pKa = 4.1DD719 pKa = 5.24LILTVNVAAA728 pKa = 5.4
MM1 pKa = 7.43VGTLKK6 pKa = 10.72DD7 pKa = 3.26IEE9 pKa = 4.43FEE11 pKa = 4.26GDD13 pKa = 2.88RR14 pKa = 11.84STRR17 pKa = 11.84DD18 pKa = 3.3LNNEE22 pKa = 3.57YY23 pKa = 11.0SDD25 pKa = 4.57AYY27 pKa = 9.47VAAVRR32 pKa = 11.84ASQANVDD39 pKa = 4.06GQGGTIRR46 pKa = 11.84GKK48 pKa = 9.58PVFTKK53 pKa = 10.91EE54 pKa = 3.49EE55 pKa = 3.57VAYY58 pKa = 9.95YY59 pKa = 10.51LNRR62 pKa = 11.84GDD64 pKa = 4.38GIIGGFEE71 pKa = 4.52SGANWDD77 pKa = 3.83GAQGYY82 pKa = 10.75ANNQYY87 pKa = 10.46YY88 pKa = 9.07WLPVTKK94 pKa = 10.68AMGGEE99 pKa = 3.87AGLPSEE105 pKa = 5.11GGPGATGPLTTLTFGFYY122 pKa = 7.63EE123 pKa = 4.4TLASLPEE130 pKa = 4.21PYY132 pKa = 10.24VYY134 pKa = 9.03RR135 pKa = 11.84TPAGNLALGLSVAQGFSPFTAAQRR159 pKa = 11.84TATRR163 pKa = 11.84EE164 pKa = 4.08AIGLWDD170 pKa = 6.15DD171 pKa = 3.95IIKK174 pKa = 10.87VNFQEE179 pKa = 4.84TSFQQGDD186 pKa = 4.26LNFMNTTTGPAQAAAYY202 pKa = 9.78LPYY205 pKa = 10.39DD206 pKa = 3.76YY207 pKa = 11.2GSEE210 pKa = 3.78EE211 pKa = 3.96GFKK214 pKa = 10.88NFEE217 pKa = 4.31GDD219 pKa = 3.37DD220 pKa = 3.46VSYY223 pKa = 11.51YY224 pKa = 10.52EE225 pKa = 3.74IAGDD229 pKa = 3.72VFVNPNQASNFQFLPGQYY247 pKa = 10.83GLNTLVHH254 pKa = 6.98EE255 pKa = 4.85IGHH258 pKa = 6.83AIGLEE263 pKa = 4.03HH264 pKa = 7.18PGAYY268 pKa = 10.25NFGPGFDD275 pKa = 3.33VTYY278 pKa = 11.0DD279 pKa = 3.31NGAEE283 pKa = 4.15YY284 pKa = 10.88YY285 pKa = 9.8QDD287 pKa = 2.89SRR289 pKa = 11.84MYY291 pKa = 10.86SIMSYY296 pKa = 9.7WDD298 pKa = 3.44AEE300 pKa = 4.43EE301 pKa = 3.96TGAAHH306 pKa = 6.68VNWEE310 pKa = 4.07NLTYY314 pKa = 10.38SYY316 pKa = 11.38NSTPMIHH323 pKa = 7.7DD324 pKa = 3.66ILAAQRR330 pKa = 11.84IYY332 pKa = 10.86GVEE335 pKa = 3.91EE336 pKa = 3.59NTRR339 pKa = 11.84TGNTTYY345 pKa = 11.09GFNSNAGKK353 pKa = 10.52DD354 pKa = 3.45MYY356 pKa = 11.4DD357 pKa = 3.23FTKK360 pKa = 10.8NPNPVISIYY369 pKa = 10.65DD370 pKa = 3.29AGGNDD375 pKa = 3.59TLDD378 pKa = 3.31LSGYY382 pKa = 6.28NTRR385 pKa = 11.84SFIDD389 pKa = 3.82LNPGAFSSAGGFYY402 pKa = 10.64SEE404 pKa = 6.3DD405 pKa = 3.43IPTLDD410 pKa = 4.11EE411 pKa = 4.14INARR415 pKa = 11.84RR416 pKa = 11.84AAAGLAPRR424 pKa = 11.84TQATYY429 pKa = 11.23DD430 pKa = 3.5LYY432 pKa = 10.77IEE434 pKa = 5.14LFGATYY440 pKa = 10.46TDD442 pKa = 3.24GLMRR446 pKa = 11.84DD447 pKa = 4.41NISIAYY453 pKa = 9.83NSIIEE458 pKa = 4.13NAVGGGGDD466 pKa = 3.67DD467 pKa = 3.62TIVANGAANRR477 pKa = 11.84IDD479 pKa = 3.83GGAGNDD485 pKa = 2.92IVSYY489 pKa = 8.05QTSGTAVSVNLANGLVSGGAAGDD512 pKa = 3.56VLISIEE518 pKa = 4.13GVTGSRR524 pKa = 11.84HH525 pKa = 5.53NDD527 pKa = 2.86VLTGSDD533 pKa = 3.75GNNVLNGGAGNDD545 pKa = 3.42IIYY548 pKa = 10.2GRR550 pKa = 11.84NGDD553 pKa = 3.77DD554 pKa = 4.28TIDD557 pKa = 3.86GGDD560 pKa = 3.88GVDD563 pKa = 3.81QLFGDD568 pKa = 4.82GGNDD572 pKa = 3.81TIRR575 pKa = 11.84GGDD578 pKa = 3.68GNDD581 pKa = 3.69TIMGGAGDD589 pKa = 5.58DD590 pKa = 3.92ILFGDD595 pKa = 4.76AGNDD599 pKa = 3.48TLYY602 pKa = 11.38GDD604 pKa = 4.75AGNDD608 pKa = 3.57VLDD611 pKa = 4.97GGAGDD616 pKa = 4.06DD617 pKa = 4.12KK618 pKa = 11.52LYY620 pKa = 11.25GGAGADD626 pKa = 3.3TFRR629 pKa = 11.84FSDD632 pKa = 3.44LGGYY636 pKa = 7.53DD637 pKa = 3.41TIVDD641 pKa = 4.24FRR643 pKa = 11.84RR644 pKa = 11.84GQGDD648 pKa = 3.92KK649 pKa = 10.48IDD651 pKa = 3.94LTGIDD656 pKa = 3.78PNINVDD662 pKa = 3.62AVNEE666 pKa = 3.94QFNFIGNAAFSGTAGEE682 pKa = 4.54LRR684 pKa = 11.84TFVDD688 pKa = 3.4RR689 pKa = 11.84GSTIIAGDD697 pKa = 3.61VNGDD701 pKa = 3.39GVADD705 pKa = 3.93FQINVGATAIRR716 pKa = 11.84ADD718 pKa = 4.1DD719 pKa = 5.24LILTVNVAAA728 pKa = 5.4
Molecular weight: 76.7 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1X7FZY8|A0A1X7FZY8_9SPHN Phosphoribosylformylglycinamidine synthase subunit PurL OS=Sphingomonas indica OX=941907 GN=purL PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.04RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84TRR21 pKa = 11.84SATVGGRR28 pKa = 11.84KK29 pKa = 9.07VLAARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.02KK41 pKa = 10.61LSAA44 pKa = 4.03
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.04RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84TRR21 pKa = 11.84SATVGGRR28 pKa = 11.84KK29 pKa = 9.07VLAARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.02KK41 pKa = 10.61LSAA44 pKa = 4.03
Molecular weight: 5.06 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
862315 |
32 |
3070 |
314.8 |
33.9 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.203 ± 0.073 | 0.719 ± 0.013 |
6.132 ± 0.037 | 5.595 ± 0.04 |
3.467 ± 0.031 | 9.095 ± 0.06 |
1.866 ± 0.024 | 4.962 ± 0.033 |
2.937 ± 0.035 | 9.756 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.346 ± 0.027 | 2.362 ± 0.03 |
5.447 ± 0.039 | 2.878 ± 0.028 |
7.71 ± 0.053 | 4.867 ± 0.034 |
5.019 ± 0.038 | 7.098 ± 0.037 |
1.377 ± 0.017 | 2.163 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
