
Faeces associated gemycircularvirus 20
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemycircularvirus; Gemycircularvirus chicas1; Chicken associated gemycircularvirus 1
Average proteome isoelectric point is 6.63
Get precalculated fractions of proteins
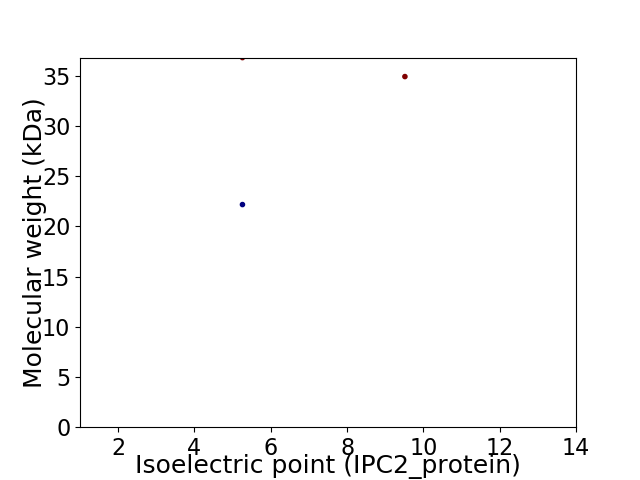
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A161J5N8|A0A161J5N8_9VIRU Replication associated protein OS=Faeces associated gemycircularvirus 20 OX=1843740 PE=3 SV=1
MM1 pKa = 7.65SAFQVNSRR9 pKa = 11.84YY10 pKa = 10.26VLLTYY15 pKa = 9.15AQCGDD20 pKa = 4.04LDD22 pKa = 3.68PWAVNDD28 pKa = 4.49LLSTLGAEE36 pKa = 4.61CIIGRR41 pKa = 11.84EE42 pKa = 3.82RR43 pKa = 11.84HH44 pKa = 5.63EE45 pKa = 5.1DD46 pKa = 3.44GGIHH50 pKa = 6.34LHH52 pKa = 6.78AFVDD56 pKa = 4.89FNRR59 pKa = 11.84KK60 pKa = 8.37FRR62 pKa = 11.84TRR64 pKa = 11.84RR65 pKa = 11.84SDD67 pKa = 2.97IFDD70 pKa = 3.44VDD72 pKa = 3.4GHH74 pKa = 6.68HH75 pKa = 7.16PNISQSRR82 pKa = 11.84GTPEE86 pKa = 3.51KK87 pKa = 10.76GYY89 pKa = 10.89DD90 pKa = 3.47YY91 pKa = 10.69AIKK94 pKa = 10.87DD95 pKa = 3.59GDD97 pKa = 4.04VVAGGLGRR105 pKa = 11.84PEE107 pKa = 4.32GKK109 pKa = 10.05SGGGDD114 pKa = 3.2GSTHH118 pKa = 6.5AKK120 pKa = 6.66WTAITQAEE128 pKa = 4.18NRR130 pKa = 11.84EE131 pKa = 4.28QFWEE135 pKa = 4.49LCHH138 pKa = 6.74EE139 pKa = 5.29LDD141 pKa = 4.04PKK143 pKa = 10.91AAATSFTQLSKK154 pKa = 9.94YY155 pKa = 10.47ADD157 pKa = 3.09WRR159 pKa = 11.84FAPDD163 pKa = 3.51PPVYY167 pKa = 9.44EE168 pKa = 4.16HH169 pKa = 7.59PIGISFTDD177 pKa = 3.42GDD179 pKa = 4.13LDD181 pKa = 5.01GRR183 pKa = 11.84RR184 pKa = 11.84EE185 pKa = 3.98WLDD188 pKa = 3.12QAGIGGGEE196 pKa = 3.44RR197 pKa = 11.84RR198 pKa = 11.84GRR200 pKa = 11.84CKK202 pKa = 10.41SLCLFGRR209 pKa = 11.84SRR211 pKa = 11.84TGKK214 pKa = 7.28TLWARR219 pKa = 11.84SLGQHH224 pKa = 6.7IYY226 pKa = 10.77CVGLVSGDD234 pKa = 3.67EE235 pKa = 4.25CMKK238 pKa = 11.14APDD241 pKa = 3.23VDD243 pKa = 3.92YY244 pKa = 11.36AIFDD248 pKa = 4.95DD249 pKa = 4.22IRR251 pKa = 11.84GGMKK255 pKa = 9.69FFPSFKK261 pKa = 9.87EE262 pKa = 3.78WLGAQAWVTVKK273 pKa = 10.56RR274 pKa = 11.84LYY276 pKa = 10.53RR277 pKa = 11.84EE278 pKa = 3.84PALVQWGKK286 pKa = 10.09PSIWLANSDD295 pKa = 3.98PRR297 pKa = 11.84NDD299 pKa = 3.35MSQDD303 pKa = 3.15DD304 pKa = 4.55VQWMEE309 pKa = 4.4DD310 pKa = 2.74NCIFVEE316 pKa = 4.32CNEE319 pKa = 4.42TIFRR323 pKa = 11.84ANIEE327 pKa = 4.03
MM1 pKa = 7.65SAFQVNSRR9 pKa = 11.84YY10 pKa = 10.26VLLTYY15 pKa = 9.15AQCGDD20 pKa = 4.04LDD22 pKa = 3.68PWAVNDD28 pKa = 4.49LLSTLGAEE36 pKa = 4.61CIIGRR41 pKa = 11.84EE42 pKa = 3.82RR43 pKa = 11.84HH44 pKa = 5.63EE45 pKa = 5.1DD46 pKa = 3.44GGIHH50 pKa = 6.34LHH52 pKa = 6.78AFVDD56 pKa = 4.89FNRR59 pKa = 11.84KK60 pKa = 8.37FRR62 pKa = 11.84TRR64 pKa = 11.84RR65 pKa = 11.84SDD67 pKa = 2.97IFDD70 pKa = 3.44VDD72 pKa = 3.4GHH74 pKa = 6.68HH75 pKa = 7.16PNISQSRR82 pKa = 11.84GTPEE86 pKa = 3.51KK87 pKa = 10.76GYY89 pKa = 10.89DD90 pKa = 3.47YY91 pKa = 10.69AIKK94 pKa = 10.87DD95 pKa = 3.59GDD97 pKa = 4.04VVAGGLGRR105 pKa = 11.84PEE107 pKa = 4.32GKK109 pKa = 10.05SGGGDD114 pKa = 3.2GSTHH118 pKa = 6.5AKK120 pKa = 6.66WTAITQAEE128 pKa = 4.18NRR130 pKa = 11.84EE131 pKa = 4.28QFWEE135 pKa = 4.49LCHH138 pKa = 6.74EE139 pKa = 5.29LDD141 pKa = 4.04PKK143 pKa = 10.91AAATSFTQLSKK154 pKa = 9.94YY155 pKa = 10.47ADD157 pKa = 3.09WRR159 pKa = 11.84FAPDD163 pKa = 3.51PPVYY167 pKa = 9.44EE168 pKa = 4.16HH169 pKa = 7.59PIGISFTDD177 pKa = 3.42GDD179 pKa = 4.13LDD181 pKa = 5.01GRR183 pKa = 11.84RR184 pKa = 11.84EE185 pKa = 3.98WLDD188 pKa = 3.12QAGIGGGEE196 pKa = 3.44RR197 pKa = 11.84RR198 pKa = 11.84GRR200 pKa = 11.84CKK202 pKa = 10.41SLCLFGRR209 pKa = 11.84SRR211 pKa = 11.84TGKK214 pKa = 7.28TLWARR219 pKa = 11.84SLGQHH224 pKa = 6.7IYY226 pKa = 10.77CVGLVSGDD234 pKa = 3.67EE235 pKa = 4.25CMKK238 pKa = 11.14APDD241 pKa = 3.23VDD243 pKa = 3.92YY244 pKa = 11.36AIFDD248 pKa = 4.95DD249 pKa = 4.22IRR251 pKa = 11.84GGMKK255 pKa = 9.69FFPSFKK261 pKa = 9.87EE262 pKa = 3.78WLGAQAWVTVKK273 pKa = 10.56RR274 pKa = 11.84LYY276 pKa = 10.53RR277 pKa = 11.84EE278 pKa = 3.84PALVQWGKK286 pKa = 10.09PSIWLANSDD295 pKa = 3.98PRR297 pKa = 11.84NDD299 pKa = 3.35MSQDD303 pKa = 3.15DD304 pKa = 4.55VQWMEE309 pKa = 4.4DD310 pKa = 2.74NCIFVEE316 pKa = 4.32CNEE319 pKa = 4.42TIFRR323 pKa = 11.84ANIEE327 pKa = 4.03
Molecular weight: 36.8 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A161IHP1|A0A161IHP1_9VIRU RepA OS=Faeces associated gemycircularvirus 20 OX=1843740 PE=3 SV=1
MM1 pKa = 7.51PARR4 pKa = 11.84YY5 pKa = 6.58PTARR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84VVRR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84PVRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84RR23 pKa = 11.84YY24 pKa = 7.32PVRR27 pKa = 11.84RR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84VFRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84STRR38 pKa = 11.84TTISRR43 pKa = 11.84RR44 pKa = 11.84AILNVASSKK53 pKa = 11.06KK54 pKa = 9.14KK55 pKa = 10.34DD56 pKa = 3.3NMVAFRR62 pKa = 11.84QTGSDD67 pKa = 3.77SEE69 pKa = 4.37PTQGPLTITGNPTLSAFGEE88 pKa = 4.67HH89 pKa = 5.59IVMWLPTARR98 pKa = 11.84NRR100 pKa = 11.84GTVPEE105 pKa = 4.08EE106 pKa = 4.06TEE108 pKa = 4.61RR109 pKa = 11.84NSQTCFMRR117 pKa = 11.84GLSEE121 pKa = 5.15RR122 pKa = 11.84INISTDD128 pKa = 2.9TSLPWTWRR136 pKa = 11.84RR137 pKa = 11.84ICFTLKK143 pKa = 9.99GLGVIRR149 pKa = 11.84RR150 pKa = 11.84DD151 pKa = 3.21GTAINVLSFEE161 pKa = 4.11EE162 pKa = 4.34AAPNGYY168 pKa = 9.27VRR170 pKa = 11.84LARR173 pKa = 11.84NYY175 pKa = 10.28SSVGGDD181 pKa = 3.81PNWEE185 pKa = 4.38AIAEE189 pKa = 4.04QLISLIMDD197 pKa = 4.02GTRR200 pKa = 11.84GIDD203 pKa = 3.95FLDD206 pKa = 3.66LTTAKK211 pKa = 9.97TDD213 pKa = 3.57GTNVTIKK220 pKa = 10.39YY221 pKa = 10.46DD222 pKa = 3.43RR223 pKa = 11.84TFRR226 pKa = 11.84LASGNDD232 pKa = 3.13SGIQKK237 pKa = 9.8VYY239 pKa = 10.95KK240 pKa = 10.0LWHH243 pKa = 6.39PMNKK247 pKa = 9.58NLVYY251 pKa = 10.83DD252 pKa = 4.4DD253 pKa = 5.8DD254 pKa = 4.96EE255 pKa = 7.3DD256 pKa = 5.7GGDD259 pKa = 3.35MVGNSISTQGKK270 pKa = 8.43PGMGDD275 pKa = 3.57YY276 pKa = 10.98YY277 pKa = 11.48VVDD280 pKa = 3.62IFKK283 pKa = 10.39AHH285 pKa = 6.95PAATSSDD292 pKa = 3.44HH293 pKa = 7.51LLFDD297 pKa = 5.29PMATLYY303 pKa = 9.48WHH305 pKa = 7.03EE306 pKa = 4.25KK307 pKa = 9.3
MM1 pKa = 7.51PARR4 pKa = 11.84YY5 pKa = 6.58PTARR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84VVRR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84PVRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84RR23 pKa = 11.84YY24 pKa = 7.32PVRR27 pKa = 11.84RR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84VFRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84STRR38 pKa = 11.84TTISRR43 pKa = 11.84RR44 pKa = 11.84AILNVASSKK53 pKa = 11.06KK54 pKa = 9.14KK55 pKa = 10.34DD56 pKa = 3.3NMVAFRR62 pKa = 11.84QTGSDD67 pKa = 3.77SEE69 pKa = 4.37PTQGPLTITGNPTLSAFGEE88 pKa = 4.67HH89 pKa = 5.59IVMWLPTARR98 pKa = 11.84NRR100 pKa = 11.84GTVPEE105 pKa = 4.08EE106 pKa = 4.06TEE108 pKa = 4.61RR109 pKa = 11.84NSQTCFMRR117 pKa = 11.84GLSEE121 pKa = 5.15RR122 pKa = 11.84INISTDD128 pKa = 2.9TSLPWTWRR136 pKa = 11.84RR137 pKa = 11.84ICFTLKK143 pKa = 9.99GLGVIRR149 pKa = 11.84RR150 pKa = 11.84DD151 pKa = 3.21GTAINVLSFEE161 pKa = 4.11EE162 pKa = 4.34AAPNGYY168 pKa = 9.27VRR170 pKa = 11.84LARR173 pKa = 11.84NYY175 pKa = 10.28SSVGGDD181 pKa = 3.81PNWEE185 pKa = 4.38AIAEE189 pKa = 4.04QLISLIMDD197 pKa = 4.02GTRR200 pKa = 11.84GIDD203 pKa = 3.95FLDD206 pKa = 3.66LTTAKK211 pKa = 9.97TDD213 pKa = 3.57GTNVTIKK220 pKa = 10.39YY221 pKa = 10.46DD222 pKa = 3.43RR223 pKa = 11.84TFRR226 pKa = 11.84LASGNDD232 pKa = 3.13SGIQKK237 pKa = 9.8VYY239 pKa = 10.95KK240 pKa = 10.0LWHH243 pKa = 6.39PMNKK247 pKa = 9.58NLVYY251 pKa = 10.83DD252 pKa = 4.4DD253 pKa = 5.8DD254 pKa = 4.96EE255 pKa = 7.3DD256 pKa = 5.7GGDD259 pKa = 3.35MVGNSISTQGKK270 pKa = 8.43PGMGDD275 pKa = 3.57YY276 pKa = 10.98YY277 pKa = 11.48VVDD280 pKa = 3.62IFKK283 pKa = 10.39AHH285 pKa = 6.95PAATSSDD292 pKa = 3.44HH293 pKa = 7.51LLFDD297 pKa = 5.29PMATLYY303 pKa = 9.48WHH305 pKa = 7.03EE306 pKa = 4.25KK307 pKa = 9.3
Molecular weight: 34.91 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
834 |
200 |
327 |
278.0 |
31.29 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.074 ± 0.387 | 1.679 ± 0.589 |
8.153 ± 0.712 | 5.156 ± 0.549 |
4.197 ± 0.449 | 9.832 ± 1.034 |
2.638 ± 0.466 | 5.276 ± 0.114 |
4.077 ± 0.108 | 6.595 ± 0.068 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.799 ± 0.501 | 3.597 ± 0.559 |
4.676 ± 0.255 | 2.998 ± 0.477 |
9.113 ± 1.324 | 6.115 ± 0.315 |
6.235 ± 1.338 | 5.156 ± 0.455 |
2.638 ± 0.396 | 2.998 ± 0.14 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
